Genetic history of Italy
The genetic history of Italy is greatly influenced by geography and history. The ancestors of Italians are mostly Indo-European speakers (Italic peoples such as Latins, Falisci, Picentes, Umbrians, Samnites, Oscans, Sicels and Adriatic Veneti, as well as Celts, Iapygians and Greeks) and pre-Indo-European speakers (Etruscans, Ligures, Rhaetians and Camunni in mainland Italy, Sicani in Sicily and the Nuragic people in Sardinia). During the Roman empire, the city of Rome also attracted people from various regions throughout the Mediterranean basin, including Southern Europe, Tunisian and the Middle East.[2] Based on DNA analysis, there is evidence of ancient regional genetic substructure and continuity within modern Italy dating to the pre-Roman and Roman periods.[3][4][5][6]
| History of Italy |
|---|
 |
|
|
In their admixture ratios, Italians are similar to other Southern Europeans, and that is being of primarily Neolithic Early European Farmer ancestry, along with smaller, but still significant, amounts of Mesolithic Western Hunter-Gatherer, Bronze Age Steppe pastoralist (Indo-European speakers) and Chalcolithic or Bronze Age Iranian/Caucasus-related ancestry.[4][7][8][9] Southern Italians are closest to the modern Greeks,[10] while the Northern Italians are closest to the Spaniards and Southern French.[11][12][13][14] There is also Bronze/Iron Age West Asian admixture in Italy, with a much lower incidence in Northern Italy compared with Central Italy and Southern Italy.[15][8] Tunisian admixture is also found in Southern Italy and the main islands, with the highest incidence being in Sicily.[15][8][4]
Overview
Latin samples from Latium in the Iron Age and early Roman Republican period were generally found to genetically cluster closest to modern Northern and Central Italians (four out of six were closest to Northern and Central Italians, while the other two were closest to Southern Italians).[16] DNA analysis demonstrates that ancient Greek colonization had a significant lasting effect on the local genetic landscape of Southern Italy and Sicily (Magna Graecia), with modern people from that region having significant Greek admixture.[17][18] Overall, the genetic differentiation between the Latins, Etruscans and the preceding proto-Villanovan population of Italy was found to be insignificant.[19] In 2019, aDNA analysis of Roman fossils detected substantial genetic ancestry shift towards Central and Northern European ancestry in the inhabitants of the city of Rome in late antiquity and the medieval era. The authors tentatively link the origin of this ancestry with Visigoths and Lombards.[2][20] A 2020 analysis of maternal haplogroups from ancient and modern samples indicates a substantial genetic similarity and continuity between the modern inhabitants of Umbria in central Italy and ancient inhabitants of the region belonging to the Italic-speaking Umbrian culture.[6]
Multiple DNA studies confirmed that genetic variation in Italy is clinal, going from the Eastern to the Western Mediterranean. The Sardinians are the exception as genetic outliers in Italy and indeed in Europe, resulting from their predominantly Neolithic, Pre-Indo-European and non-Italic Nuragic ancestry.[21][7][8][9] Reflecting the history of Europe and the broader Mediterranean basin, the Italian populations have been found to be made up mostly of the same ancestral components, albeit in different proportions, from the Mesolithic, Neolithic and Bronze Age settlements of Europe.[15][12][22]
The genetic gap between northern and southern Italians is filled by an intermediate Central Italian cluster, creating a continuous cline of variation that mirrors geography.[23] The only exceptions are some minority populations (mostly Slovene minorities from the region of Friuli-Venezia Giulia) who cluster with the Slavic-speaking Central Europeans in Slovenia,[24] as well as the Sardinians, who are clearly differentiated from the populations of both mainland Italy and Sicily.[12][4] A study on some linguistic and isolated communities residing in Italy revealed that their genetic diversity at short (0–200 km) and intermediate distances (700–800 km) was greater than that observed throughout the entire European continent.[5]
The genetic distance between Northern and Southern Italians, although large for a single European nationality, is similar to that between the Northern and the Southern Germans.[25] Northern and Southern Italians began to diverge as early as the Late Glacial, and appear to encapsulate at a smaller scale the cline of genetic diversity observable across Europe.[26]
Historical populations of Italy
Modern humans appeared during the Upper Paleolithic. Specimens of Aurignacian age were discovered in the cave of Fumane and date back about 34,000 years. During the Magdalenian period, the first humans from the Pyrenees populated Sardinia.[27]
During the Neolithic period, farming was introduced by people from the east and the first villages were built. Weapons became more sophisticated and the first objects in clay were produced. In the late Neolithic era the use of copper spread, and villages were built over piles near lakes. In Sardinia, Sicily and a part of Mainland Italy the Beaker culture spread from Western and Central Europe.
During the Late Bronze Age the Urnfield Proto-Villanovan culture appeared in Central and Northern Italy. It is characterized by the rite of cremation of dead bodies, which originated from Central Europe. The use of iron began to spread.[28] In Sardinia, the Nuragic civilization flourished.
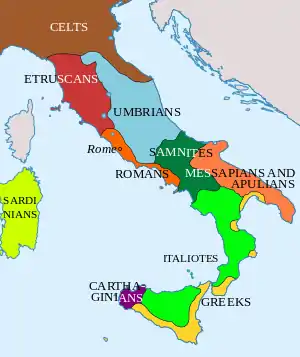
At the dawn of the Iron Age much of Italy was inhabited by Italic tribes such as the Latins, Sabines, Samnites, and Umbrians. The Northwest and Alpine territories were populated primarily by pre-Indo European speakers such as the Etruscans, Ligurians, Camunni and Raetians; while Iapygian tribes, possibly of Illyrian origin, populated Apulia.
From the 8th century BC, Greek colonists settled on the southern Italian coast and founded cities, forming what would be later called Magna Graecia. Around the same time, Phoenician colonists settled on the western side of Sicily. During the same period the Etruscan civilization developed on the coast of Southern Tuscany and Northern Latium. In the 4th century BC, Gauls settled in Northern Italy and in parts of Central Italy.[29] With the fall of the Western Roman Empire, different populations of Germanic origin invaded Italy, the most significant being the Lombards,[30] followed five centuries later by the Normans in Sicily.
Y-DNA genetic diversity

Many Italians, especially in Northern Italy and Central Italy, belong to Haplogroup R1b, common in Western and Central Europe. The highest frequency of R1b is found in Garfagnana (76.2%) in Tuscany and in the Bergamo Valleys (80.8%) in Lombardy.[31][32] This percentage lowers in the south of Italy in Calabria (33.2%).[31] On the other hand 39% of the Sardinians belong to Mesolithic European haplogroup I2a1a.[33][34]
A study from the Università Cattolica del Sacro Cuore found that while Greek colonization left little significant genetic contribution, data analysis sampling 12 sites in the Italian peninsula supported a male demic diffusion model and Neolithic admixture with Mesolithic inhabitants.[35] The results supported a distribution of genetic variation along a north–south axis and supported demic diffusion. South Italian samples clustered with southeast and south-central European samples, and northern groups with West Europe.[35][36]
A 2004 study by Semino et al. showed that Italians from the north-central regions had around 26.9% J2; the Apulians, Calabrians and Sicilians had 29.1%, 21.5% and 16.7% J2 respectively; the Sardinians had 9.7% J2.[37]
A 2018 genetic study, focusing on the Y-chromosome and haplogroups lineages, their diversity and their distribution by taking some 817 representative subjects, gives credit to the traditional northern-southern division in population, by concluding that due to Neolithic migrations southern Italians "show a higher similarity with Middle Eastern and Southern Balkan populations than northern ones; conversely, northern samples are genetically closer to North-West Europe and Northern Balkan groups". The position of Volterra in central Tuscany keeps the debate about the origins of Etruscans open, although the numbers are strongly in favor of the autochthonous thesis: the low presence of J2a-M67* (2.7%) suggests contacts by sea with Anatolian people; the presence of Central European lineage G2a-L497 (7.1%) at considerable frequency would rather support a Central European origin of the Etruscans; and finally, the high incidence of European R1b lineages (R1b 50% approx., R1b-U152 24.5%) — especially of haplogroup R1b-U152 — could suggest an autochthonous origin due to a process of formation of the Etruscan civilisation from the preceding Villanovan culture, following the theories of Dionysius of Halicarnassus,[31] as already supported by archaeology, anthropology and linguistics.[38][39][40][41] In 2019, in a Stanford study published in Science, two ancient samples from the Neolithic settlement of Ripabianca di Monterado in province of Ancona, in the Marche region of Italy, were found to be Y-DNA J-L26 and J-M304.[2] Therefore, Y-DNA J2a-M67, downstream to Y-DNA J-L26 and J-M304, is most likely in Italy since the Neolithic and can't be the proof of recent contacts with Anatolia.
Y-DNA introduced by historical immigration
In two villages in Lazio and Abruzzo (Cappadocia and Vallepietra), I1 is the most common Y-DNA, recorded at levels 35% and 28%.[42] In Sicily, further migrations from the Vandals and Saracens have only slightly affected the ethnic composition of the Sicilian people. However, specifically Greek genetic legacy is estimated at 37% in Sicily.[17]
The Norman conquest of southern Italy caused the Norman Kingdom of Sicily to be created in 1130, with Palermo as capital, 70 years after the initial Norman invasion and 40 after the conquest of the last town, Noto in 1091, and would last until 1198. Nowadays it is in north-west Sicily, around Palermo and Trapani, that Norman Y-DNA is common, with 15% to 20% of the lineages belonging to haplogroup I. The North African male contribution to Sicily was estimated between 0% and 7.5%.[43][18][44] Overall, the estimated Southern Balkan and Western European paternal contributions in Sicily are about 63% and 26% respectively.[44]
A 2015 genetic study of six small mountain villages in eastern Lazio and one mountain community in nearby western Abruzzo found some genetic similarities between these communities and Near Eastern populations, mainly in the male genetic pool. The Y haplogroup Q, common in Western Asia and Central Asia, was also found among this sample population, suggesting that in the past could have hosted a settlement from Central Asia.[42] Also, it is about 0.6% in continental Italy, but it rises to 2.5% (6/236) in Sicily, where it reaches 16.7% (3/18) in Mazara del Vallo region, followed by 7.1% (2/28) in Ragusa, 3.6% in Sciacca,[17] and 3.7% in Belvedere Marittimo.[45]
Genetic composition of Italian mtDNA
In Italy as elsewhere in Europe the majority of mtDNA lineages belong to the haplogroup H. Several independent studies conclude that haplogroup H probably evolved in West Asia c. 25,000 years ago. It was carried to Europe by migrations c. 20–25,000 years ago, and spread with population of the southwest of the continent.[46][47] Its arrival was roughly contemporary with the rise of the Gravettian culture. The spread of subclades H1, H3 and the sister haplogroup V reflect a second intra-European expansion from the Franco-Cantabrian region after the last glacial maximum, c. 13,000 years ago.[46][48]
African Haplogroup L lineages are relatively infrequent (less than 1%) throughout Italy with the exception of Latium, Volterra, Basilicata and Sicily where frequencies between 2 and 3% have been found.[49]
A study in 2012 by Brisighelli et al. stated that an analysis of ancestral informative markers "as carried out in the present study indicated that Italy shows a very minor sub-Saharan African component that is, however, slightly higher than non-Mediterranean Europe." Discussing African mtDNAs the study states that these indicate that a significant proportion of these lineages could have arrived in Italy more than 10,000 years ago; therefore, their presence in Italy does not necessarily date to the time of the Roman Empire, the Atlantic slave trade or to modern migration."[22] These mtDNAs by Brisighelli et al. were reported with the given results as "Mitochondrial DNA haplotypes of African origin are mainly represented by haplogroups M1 (0.3%), U6 (0.8%) and L (1.2%) for the 583 samples tested.[22] The haplogroups M1 and U6 can be considered to be of North African origin and could therefore be used to signal the documented African historical input. Haplogroup M1 was observed in only two carriers from Trapani (West Sicily), while U6 was observed only in Lucera, South Apulia, and another at the tip of the Peninsula (Calabria).[22]
A 2013 study by Alessio Boattini et al. found 0 of African L haplogroup in the whole Italy out of 865 samples. The percentages for Berber M1 and U6 haplogroups were 0.46% and 0.35%, respectively.[50]
A 2014 study by Stefania Sarno et al. found 0 of African L and M1 haplogroups in mainland Southern Italy out of 115 samples. Only two Berber U6 out of 115 samples were found, one from Lecce and one from Cosenza.[44]
A close genetic similarity between Ashkenazim and Italians has been noted in genetic studies, possibly due to the fact that Ashkenazi Jews have a significant European admixture (30–60%), much of it Southern European, a lot of which came from Italy when Jewish diaspora males of Middle Eastern origin migrated to Rome and found wives among local women who then converted to Judaism.[51][52][53][54][55][13][56] More specifically, Ashkenazi Jews could be modeled as being 50% Levantine and 50% European, with an estimated mean South European admixture of 37.5%. Most of it (30.5%) seems to derive from an Italian source.[57][58]
A 2010 study of Jewish genealogy found that with respect to non-Jewish European groups, the populations which are most closely related to Ashkenazi Jews are modern-day Italians followed by the French and Sardinians.[59][60]
Recent studies have shown that Italy played an important role in the recovery of "Western Europe" at the end of the Last glacial period. The study which was focused on the mitochondrial U5b3 haplogroup discovered that this female lineage had in fact originated in Italy and around 10,000 years ago it expanded from the Peninsula towards Provence and the Balkans. In Provence, probably between 9,000 and 7,000 years ago, it gave rise to the haplogroup subclade U5b3a1. This subclade U5b3a1 later came from Provence to the island of Sardinia by way of obsidian merchants, because it is estimated that 80% of the obsidian which is found in France comes from Monte Arci in Sardinia, reflecting the close relationship which once existed between these two regions. Still about 4% of the female population of Sardinia belongs to this haplotype.[61]
A mtDNA study, published in 2018 in the journal American Journal of Physical Anthropology, compared both ancient and modern samples from Tuscany, from the Prehistory, Etruscan age, Roman age, Renaissance, and Present-day, and concluded that the Etruscans appear as a local population, intermediate between the prehistoric and the other samples, placing in the temporal network between the Eneolithic Age and the Roman Age.[62]
A 2020 analysis of maternal haplogroups from ancient and modern samples in the central Italian region of Umbria finds a substantial genetic similarity among modern Umbrians and the area's pre-Roman inhabitants, and evidence of substantial genetic continuity in the region from pre-Roman times to the present. Both modern and ancient Umbrians were found to have high rates of mtDNA haplogroups U4 and U5a, and an overrepresentation of J (at roughly 30%). The study also found that, "local genetic continuities are further attested to by six terminal branches (H1e1, J1c3, J2b1, U2e2a, U8b1b1 and K1a4a)" also shared by ancient and modern Umbrians.[6]
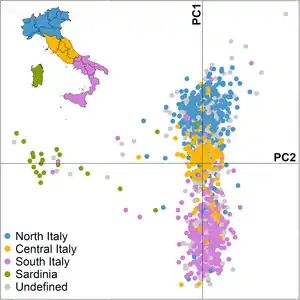
Autosomal

South/West European North/East European Caucasus
West Asian South Asian East Asian North African/Sub-Saharan African[63]
_PC_analysis.png.webp)
Wade et al. (2008) determined that Italy is one of the last two remaining genetic islands in Europe, the other being Finland. This is due in part to the presence of the Alpine mountain chain which, over the centuries, has prevented large migration flows. [64]
Recent genome-wide studies have been able to detect and quantify admixture like never before. Li et al. (2008), using more than 600,000 autosomal SNPs, identify seven global population clusters, including European, Middle Eastern and Central/South Asian. All the Italian samples belong to Central-Western group with minor influences dating to Neolithic period.[65]
López Herráez et al. (2009) typed the same samples at close to 1 million SNPs and analyzed them in a Western Eurasian context, identifying a number of subclusters. This time, all of the European samples show some minor admixture. Among the Italians, Tuscany still has the most, and Sardinia has a bit too, but so does Lombardy (Bergamo), which is even farther north.[66]
A 2011 study by Moorjani et al. found that many southern Europeans have inherited 1–3% Sub-Saharan ancestry, although the percentages were lower (0.2–2.1%) when reanalyzed with the 'STRUCTURE' statistical model. An average admixture date of around 55 generations/1100 years ago was also calculated, "consistent with North African gene flow at the end of the Roman Empire and subsequent Arab migrations"[67]
A 2012 study by Di Gaetano et al. used 1,014 Italians with wide geographical coverage. It showed that the current population of Sardinia can be clearly differentiated genetically from mainland Italy and Sicily, and that a certain degree of genetic differentiation is detectable within the current Italian peninsula population.
By using the ADMIXTURE software, the authors obtained at K = 4 the lowest cross-validation error. The HapMap CEU individuals showed an average Northern Europe (NE) ancestry of 83%. A similar pattern is observed in French, Northern Italian and Central Italian populations with a NE ancestry of 70%, 56% and 52% respectively. According to the PCA plot, also in the ADMIXTURE analysis there are relatively small differences in ancestry between Northern Italians and Central Italians while Southern Italians showed a lower average admixture NE proportion (44%) than Northern and Central Italy, and a higher Caucasian ancestry of 28%. The Sardinian samples display a pattern of crimson common to the others European populations but at a higher frequency (70%). The average admixture proportions for Northern European ancestry within current Sardinian population is 14.3% with some individuals exhibiting very low Northern European ancestry (less than 5% in 36 individuals on 268 accounting the 13% of the sample).[12]
A 2013 study by Peristera Paschou et al. confirms that the Mediterranean Sea has acted as a strong barrier to gene flow through geographic isolation following initial settlements. Samples from (Northern) Italy, Tuscany, Sicily and Sardinia are closest to other Southern Europeans from Iberia, the Balkans and Greece, who are in turn closest to the Neolithic migrants that spread farming throughout Europe, represented here by the Cappadocian sample from Anatolia. But there hasn't been any significant admixture from the Middle East or North Africa into Italy and the rest of Southern Europe since then.[14]
Ancient DNA analysis reveals that Ötzi the Iceman clusters with modern Southern Europeans and closest to Italians (the orange "Europe S" dots in the plots below), especially those from the island of Sardinia. Other Italians pull away toward Southeastern and Central Europe consistent with geography and some post-Neolithic gene flow from those areas (e.g. Italics, Greeks, Etruscans, Celts), but despite that and centuries of history, they're still very similar to their prehistoric ancestor.[68]
A 2013 study by Botigué et al. 2013 applied an unsupervised clustering algorithm, ADMIXTURE, to estimate allele-based sharing between Africans and Europeans. Regarding Italians, the North African ancestry does not exceed 2% of their genomes. On average, 1% of Jewish ancestry is found in Tuscan HapMap population and Italian Swiss, as well as Greeks and Cypriots. Contrary to past observations, Sub-Saharan ancestry is detected at <1% in Europe, with the exception of the Canary Islands.[44]
Haak et al. (2015) conducted a genome wide study of 94 ancient skeletons from Europe and Russia. The study argues that Bronze Age steppe pastoralists from the Yamna culture spread Indo-European languages in Europe. Autosomic tests indicate that the Yamnaya-people were the result of admixture between two different hunter-gatherer populations: Eastern Hunter-Gatherers from the Russian Steppe and either Caucasus Hunter-Gatherers or Chalcolithic Iranians (who are very similar). Wolfgang Haak estimated a 27% ancestral contribution of the Yamnaya in the DNA of modern Tuscans, a 25% ancestral contribution of the Yamnaya in the DNA of modern Northern Italians from Bergamo, excluding Sardinians (7%), and to a lesser extent Sicilians (12%).[11]
A 2016 study Sazzini et al., confirms the results of previous studies by Di Gaetano et al. (2012) and Fiorito et al. (2015) but has much better geographical coverage of samples, with 737 individuals from 20 locations in 15 different regions being tested. The study also for the first time includes a formal admixture test that models the ancestry of Italians by inferring admixture events using all of the Western Eurasian samples. The results are very interesting in light of the ancient DNA evidence that has come out in the last couple years:
In addition to the pattern described in the main text, the SARD sample seemed to have played a major role as source of admixture for most of the examined populations, especially Italian ones, rather than as recipient of migratory processes. In fact, the most significant f3 scores for trios including SARD indicated peninsular Italians as plausible results of admixture between SARD and populations from Iran, Caucasus and Russia. This scenario could be interpreted as further evidence that Sardinians retain high proportions of a putative ancestral genomic background that was considerably widespread across Europe at least until the Neolithic and that has been subsequently erased or masked in most of present-day European populations.[23]
Sarno et al. (2017) concentrate on the genetic impact brought by the historical migrations around the Mediterranean on Southern Italy and Sicily, and conclude that the "results demonstrate that the genetic variability of present-day Southern Italian populations is characterized by a shared genetic continuity, extending to large portions of central and eastern Mediterranean shores", while showing that "Southern Italy appear more similar to the Greek-speaking islands of the Mediterranean Sea, reaching as far east as Cyprus, than to samples from continental Greece, suggesting a possible ancestral link which might have survived in a less admixed form in the islands", also precises how "besides a predominant Neolithic-like component, our analyses reveal significant impacts of Post-Neolithic Caucasus- and Levantine-related ancestries."[18] A news article associated with the Max Planck Society, reviewing the results, while beginning by stating that "populations along the eastern Mediterranean coast share a genetic heritage that transcends nationality", also points out how this study is interesting on the debates concerning the diffusion of the Indo-European languages family in Europe, as, while showcasing the influence from the Caucasus, there's no genetic marker associated with the Pontic–Caspian steppe, "a very characteristic genetic signal well represented in North-Central and Eastern Europe, which previous studies associated with the introduction of Indo-European languages to the continent."[18]
Raveane et al. (2019) discovered in a genome-wide study on modern-day Italians a contribution of Caucasus Hunter-Gatherers from the third millennium Anatolian Bronze Age, predominantly in Southern Italy. Furthermore, patterns of regional variation showed geographical structure in Southern Italy, Northern Italy, and Sardinia, in line with previous studies. Even more detailed structure was observed between subregional clusters, caused by geography and distance, and historical admixture possibly associated with events at the end of the Roman Empire and during subsequent periods.[4]
Antonio et al. (2019) studied historical populations from various time periods in Latium and Rome. They found that, despite the linguistic differences, the Latins and the Etruscans showed no significant genetic differences. Their autosomal DNA was a mixture in similar proportions of Western Hunter-Gatherers (Mesolithic), Early European Farmers (Neolithic), and Western Steppe Herders (Bronze Age).[2]
A 2022 genome-wide study of more than 700 individuals from the South Mediterranean area (102 from Southern Italy), combined with ancient DNA from neighbouring areas, found high affinities of South-Eastern Italians with modern Eastern Peloponnesians, and a closer affinity of ancient Greek genomes with those from specific regions of South Italy than modern Greek genomes.[69]
References
- Parolo S, Lisa A, Gentilini D, Di Blasio AM, Barlera S, Nicolis EB, et al. (November 2015). "Characterization of the biological processes shaping the genetic structure of the Italian population". BMC Genetics. 16: 132. doi:10.1186/s12863-015-0293-x. PMC 4640365. PMID 26553317.
- Antonio ML, Gao Z, Moots HM, Lucci M, Candilio F, Sawyer S, et al. (November 2019). "Ancient Rome: A genetic crossroads of Europe and the Mediterranean". Science. American Association for the Advancement of Science (published November 8, 2019). 366 (6466): 708–714. Bibcode:2019Sci...366..708A. doi:10.1126/science.aay6826. hdl:2318/1715466. PMC 7093155. PMID 31699931.
Interestingly, although Iron Age individuals were sampled from both Etruscan (n=3) and Latin (n=6) contexts, we did not detect any significant differences between the two groups with f4 statistics in the form of f4(RMPR_Etruscan, RMPR_Latin; test population, Onge), suggesting shared origins or extensive genetic exchange between them. ... In the Medieval and early modern periods (n = 28 individuals), we observe an ancestry shift toward central and northern Europe in PCA (Fig. 3E), as well as a further increase in the European cluster (C7) and loss of the Near Eastern and eastern Mediterranean clusters (C4 and C5) in ChromoPainter (Fig. 4C). The Medieval population is roughly centered on modern-day central Italians (Fig. 3F). It can be modeled as a two-way combination of Rome's Late Antique population and a European donor population, with potential sources including many ancient and modern populations in central and northern Europe: Lombards from Hungary, Saxons from England, and Vikings from Sweden, among others (table S26).
- Ralph P, Coop G (2013). "The geography of recent genetic ancestry across Europe". PLOS Biology. 11 (5): e1001555. doi:10.1371/journal.pbio.1001555. PMC 3646727. PMID 23667324.
- Raveane A, Aneli S, Montinaro F, Athanasiadis G, Barlera S, Birolo G, et al. (September 2019). "Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe". Science Advances. 5 (9): eaaw3492. Bibcode:2019SciA....5.3492R. doi:10.1126/sciadv.aaw3492. PMC 6726452. PMID 31517044.
- Capocasa M, Anagnostou P, Bachis V, Battaggia C, Bertoncini S, Biondi G, et al. (2014). "Linguistic, geographic and genetic isolation: a collaborative study of Italian populations". Journal of Anthropological Sciences. 92 (92): 201–31. doi:10.4436/JASS.92001. PMID 24607994.
- Modi A, Lancioni H, Cardinali I, Capodiferro MR, Rambaldi Migliore N, Hussein A, et al. (July 2020). "The mitogenome portrait of Umbria in Central Italy as depicted by contemporary inhabitants and pre-Roman remains". Scientific Reports. 10 (1): 10700. Bibcode:2020NatSR..1010700M. doi:10.1038/s41598-020-67445-0. PMC 7329865. PMID 32612271.
- Chiang CW, Marcus JH, Sidore C, Biddanda A, Al-Asadi H, Zoledziewska M, et al. (October 2018). "Genomic history of the Sardinian population". Nature Genetics. 50 (10): 1426–1434. doi:10.1038/s41588-018-0215-8. PMC 6168346. PMID 30224645.
- Marcus JH, Posth C, Ringbauer H, Lai L, Skeates R, Sidore C, et al. (February 2020). "Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia". Nature Communications. 11 (1): 939. Bibcode:2020NatCo..11..939M. doi:10.1038/s41467-020-14523-6. PMC 7039977. PMID 32094358.
- Fernandes DM, Mittnik A, Olalde I, Lazaridis I, Cheronet O, Rohland N, et al. (March 2020). "The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean". Nature Ecology & Evolution. 4 (3): 334–345. doi:10.1038/s41559-020-1102-0. PMC 7080320. PMID 32094539.
- «Sicily and Southern Italy were heavily colonized by Greeks beginning in the eight to ninth century B.C.. The demographic development of the Greek colonies in Southern Italy was remarkable, and in classical times this region was called Magna Graecia (Great Greece) because it probably surpassed in numbers the Greek population of the motherland.» Cavalli-Sforza L, Menozzi P, Piazza A (1994). The History and Geography of Human Genes. Princeton University Press. p. 278. ISBN 978-0-691-08750-4.
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, et al. (June 2015). "Massive migration from the steppe was a source for Indo-European languages in Europe". Nature. 522 (7555): 207–11. arXiv:1502.02783. Bibcode:2015Natur.522..207H. doi:10.1038/nature14317. PMC 5048219. PMID 25731166.
- Di Gaetano C, Voglino F, Guarrera S, Fiorito G, Rosa F, Di Blasio AM, et al. (2012). "An overview of the genetic structure within the Italian population from genome-wide data". PLOS ONE. 7 (9): e43759. Bibcode:2012PLoSO...743759D. doi:10.1371/journal.pone.0043759. PMC 3440425. PMID 22984441.
- Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, et al. (January 2008). "Discerning the ancestry of European Americans in genetic association studies". PLOS Genetics. 4 (1): e236. doi:10.1371/journal.pgen.0030236. PMC 2211542. PMID 18208327.
- Paschou P, Drineas P, Yannaki E, Razou A, Kanaki K, Tsetsos F, et al. (June 2014). "Maritime route of colonization of Europe". Proceedings of the National Academy of Sciences of the United States of America. 111 (25): 9211–6. Bibcode:2014PNAS..111.9211P. doi:10.1073/pnas.1320811111. PMC 4078858. PMID 24927591.
- Fiorito G, Di Gaetano C, Guarrera S, Rosa F, Feldman MW, Piazza A, Matullo G (July 2016). "The Italian genome reflects the history of Europe and the Mediterranean basin". European Journal of Human Genetics. 24 (7): 1056–62. doi:10.1038/ejhg.2015.233. PMC 5070887. PMID 26554880.
- Antonio et al. 2019, p. 2.
- Di Gaetano C, Cerutti N, Crobu F, Robino C, Inturri S, Gino S, et al. (January 2009). "Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome". European Journal of Human Genetics. 17 (1): 91–9. doi:10.1038/ejhg.2008.120. PMC 2985948. PMID 18685561.
The genetic contribution of Greek chromosomes to the Sicilian gene pool is estimated to be about 37% whereas the contribution of North African populations is estimated to be around 6%.
- Sarno S, Boattini A, Pagani L, Sazzini M, De Fanti S, Quagliariello A, et al. (May 2017). "Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean". Scientific Reports. 7 (1): 1984. Bibcode:2017NatSR...7.1984S. doi:10.1038/s41598-017-01802-4. PMC 5434004. PMID 28512355.
- Antonio et al. 2019, p. 3.
- Wade, Lizzie (2019). "Immigrants from the Middle East shaped Rome". Science. 366 (6466): 673. Bibcode:2019Sci...366..673W. doi:10.1126/science.366.6466.673. PMID 31699914. S2CID 207965960. Archived from the original on 2021-02-24. Retrieved 2021-01-25."Trade routes sent people and goods to the new capital, and epidemics and invasions reduced Rome's population to about 100,000 people. Invading barbarians brought in more European ancestry. Rome gradually lost its strong genetic link to the Eastern Mediterranean and Middle East. By medieval times, city residents again genetically resembled European populations."
- ..."La separazione della Sardegna dal resto del continente, anzi da tutte le altre popolazioni europee, che probabilmente rivela un'origine più antica della sua popolazione, indipendente da quella delle popolazioni italiche e con ascendenze nel Mediterraneo Medio-Orientale." Alberto Piazza, I profili genetici degli italiani, Accademia delle Scienze di Torino
- Brisighelli F, Álvarez-Iglesias V, Fondevila M, Blanco-Verea A, Carracedo A, Pascali VL, et al. (2012). "Uniparental markers of contemporary Italian population reveals details on its pre-Roman heritage". PLOS ONE. 7 (12): e50794. Bibcode:2012PLoSO...750794B. doi:10.1371/journal.pone.0050794. PMC 3519480. PMID 23251386.
- Sazzini M, Gnecchi Ruscone GA, Giuliani C, Sarno S, Quagliariello A, De Fanti S, et al. (September 2016). "Complex interplay between neutral and adaptive evolution shaped differential genomic background and disease susceptibility along the Italian peninsula". Scientific Reports. 6: 32513. Bibcode:2016NatSR...632513S. doi:10.1038/srep32513. PMC 5007512. PMID 27582244. Zendo: 165505.
- Esko T, Mezzavilla M, Nelis M, Borel C, Debniak T, Jakkula E, et al. (June 2013). "Genetic characterization of northeastern Italian population isolates in the context of broader European genetic diversity". European Journal of Human Genetics. 21 (6): 659–65. doi:10.1038/ejhg.2012.229. PMC 3658181. PMID 23249956.
- Nelis M, Esko T, Mägi R, Zimprich F, Zimprich A, Toncheva D, et al. (2009). "Genetic structure of Europeans: a view from the North-East". PLOS ONE. 4 (5): e5472. Bibcode:2009PLoSO...4.5472N. doi:10.1371/journal.pone.0005472. PMC 2675054. PMID 19424496.
- Sazzini M, Abondio P, Sarno S, Gnecchi-Ruscone GA, Ragno M, Giuliani C, et al. (May 2020). "Genomic history of the Italian population recapitulates key evolutionary dynamics of both Continental and Southern Europeans". BMC Biology. 18 (1): 51. doi:10.1186/s12915-020-00778-4. PMC 7243322. PMID 32438927.
- Rootsi S (December 2006). "Y-Chromosome haplogroup I prehistoric gene flow in Europe" (PDF). Documenta Praehistorica. 33: 17–20. doi:10.4312/dp.33.3. Archived from the original (PDF) on 2009-03-06.
- "Culture del bronzo recente in Italia settentrionale e loro rapporti con la "cultura dei campi di urne"". Gruppo Archeologico Monteclarense. Archived from the original on 10 May 2006.
- Haywood J (2014). The Celts: Bronze Age to New Age. Routledge. p. 21. ISBN 978-1-317-87017-3.
- "Lombard people". Encyclopaedia Britannica.
- Grugni V, Raveane A, Mattioli F, Battaglia V, Sala C, Toniolo D, et al. (February 2018). "Reconstructing the genetic history of Italians: new insights from a male (Y-chromosome) perspective". Annals of Human Biology. 45 (1): 44–56. doi:10.1080/03014460.2017.1409801. PMID 29382284. S2CID 43501209.
- Di Giacomo F, Luca F, Anagnou N, Ciavarella G, Corbo RM, Cresta M, et al. (September 2003). "Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects" (PDF). Molecular Phylogenetics and Evolution. 28 (3): 387–95. doi:10.1016/s1055-7903(03)00016-2. PMID 12927125. Archived from the original (PDF) on 2017-01-20.
- Francalacci, P.; Morelli, L.; Underhill, P. A.; Lillie, A. S.; Passarino, G.; Useli, A.; Madeddu, R.; Paoli, G.; Tofanelli, S.; Calò, C. M.; Ghiani, M. E.; Varesi, L.; Memmi, M.; Vona, G.; Lin, A. A. (July 2003). "Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability". American Journal of Physical Anthropology. 121 (3): 270–279. doi:10.1002/ajpa.10265. ISSN 0002-9483. PMID 12772214.
- Francalacci, Paolo; Morelli, Laura; Angius, Andrea; Berutti, Riccardo; Reinier, Frederic; Atzeni, Rossano; Pilu, Rosella; Busonero, Fabio; Maschio, Andrea; Zara, Ilenia; Sanna, Daria; Useli, Antonella; Urru, Maria Francesca; Marcelli, Marco; Cusano, Roberto (2013-08-02). "Low-Pass DNA Sequencing of 1200 Sardinians Reconstructs European Y-Chromosome Phylogeny". Science. 341 (6145): 565–569. Bibcode:2013Sci...341..565F. doi:10.1126/science.1237947. ISSN 0036-8075. PMC 5500864. PMID 23908240.
- Capelli C, Brisighelli F, Scarnicci F, Arredi B, Caglia' A, Vetrugno G, et al. (July 2007). "Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter". Molecular Phylogenetics and Evolution. 44 (1): 228–39. doi:10.1016/j.ympev.2006.11.030. PMID 17275346.
- Cavalli-Sforza L, Menozzi P, Piazza A (1994). The History and Geography of Human Genes. Princeton University Press. ISBN 978-0-691-08750-4.>: 295
- Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, et al. (May 2004). "Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area". American Journal of Human Genetics. 74 (5): 1023–34. doi:10.1086/386295. PMC 1181965. PMID 15069642.
- Barker, Graeme; Rasmussen, Tom (2000). The Etruscans. The Peoples of Europe. Oxford: Blackwell Publishing. p. 44. ISBN 978-0-631-22038-1.
- Turfa, Jean MacIntosh (2017). "The Etruscans". In Farney, Gary D.; Bradley, Gary (eds.). The Peoples of Ancient Italy. Berlin: De Gruyter. pp. 637–672. doi:10.1515/9781614513001. ISBN 978-1-61451-520-3.
- De Grummond, Nancy T. (2014). "Ethnicity and the Etruscans". In McInerney, Jeremy (ed.). A Companion to Ethnicity in the Ancient Mediterranean. Chichester, UK: John Wiley & Sons, Inc. pp. 405–422. doi:10.1002/9781118834312. ISBN 9781444337341.
- Shipley, Lucy (2017). "Where is home?". The Etruscans: Lost Civilizations. London: Reaktion Books. pp. 28–46. ISBN 9781780238623.
- Messina F, Finocchio A, Rolfo MF, De Angelis F, Rapone C, Coletta M, et al. (2015). "Traces of forgotten historical events in mountain communities in Central Italy: A genetic insight". American Journal of Human Biology. 27 (4): 508–19. doi:10.1002/ajhb.22677. PMID 25728801. S2CID 30111156.
- Capelli C, Onofri V, Brisighelli F, Boschi I, Scarnicci F, Masullo M, et al. (June 2009). "Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe". European Journal of Human Genetics. 17 (6): 848–52. doi:10.1038/ejhg.2008.258. PMC 2947089. PMID 19156170.
- Sarno S, Boattini A, Carta M, Ferri G, Alù M, Yao DY, et al. (2014). "An ancient Mediterranean melting pot: investigating the uniparental genetic structure and population history of sicily and southern Italy". PLOS ONE. 9 (4): e96074. Bibcode:2014PLoSO...996074S. doi:10.1371/journal.pone.0096074. PMC 4005757. PMID 24788788.
 This article contains quotations from this source, which is available under a Creative Commons Attribution 4.0 International (CC BY 4.0) license.
This article contains quotations from this source, which is available under a Creative Commons Attribution 4.0 International (CC BY 4.0) license. - Brisighelli, F (2012). "Uniparental markers of contemporary Italian population reveals details on its pre-Roman heritage". PLOS ONE. 7 (12): e50794. Bibcode:2012PLoSO...750794B. doi:10.1371/journal.pone.0050794. PMC 3519480. PMID 23251386. In Table S4, #BEL50 is estimated to be Q-M378 by haplotype, though it is shown as just P* (xR1).
- Pereira L, Richards M, Goios A, Alonso A, Albarrán C, Garcia O, et al. (January 2005). "High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium". Genome Research. 15 (1): 19–24. doi:10.1101/gr.3182305. PMC 540273. PMID 15632086.
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. (November 2000). "Tracing European founder lineages in the Near Eastern mtDNA pool". American Journal of Human Genetics. 67 (5): 1251–76. doi:10.1016/S0002-9297(07)62954-1. PMC 1288566. PMID 11032788.
- Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, Scozzari R, et al. (November 2004). "The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool". American Journal of Human Genetics. 75 (5): 910–8. doi:10.1086/425590. PMC 1182122. PMID 15382008.
- Achilli A, Olivieri A, Pala M, Metspalu E, Fornarino S, Battaglia V, et al. (April 2007). "Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans". American Journal of Human Genetics. 80 (4): 759–68. doi:10.1086/512822. PMC 1852723. PMID 17357081.
4/138=2.9% in Latium; 3/114=2.6% in Volterra; 2/92=2.2% in Basilicata and 3/154=2.0% in Sicily
- Boattini A, Martinez-Cruz B, Sarno S, Harmant C, Useli A, Sanz P, et al. (2013). "Uniparental markers in Italy reveal a sex-biased genetic structure and different historical strata". PLOS ONE. 8 (5): e65441. Bibcode:2013PLoSO...865441B. doi:10.1371/journal.pone.0065441. PMC 3666984. PMID 23734255.
- Balter M (3 June 2010). "Tracing the Roots of Jewishness". Science Magazine. American Association for the Advancement of Science.
- Zoossmann-Diskin A (October 2010). "The origin of Eastern European Jews revealed by autosomal, sex chromosomal and mtDNA polymorphisms". Biology Direct. 5 (57): 57. doi:10.1186/1745-6150-5-57. PMC 2964539. PMID 20925954.
- Balter M (8 October 2013). "Did Modern Jews Originate in Italy?". ScienceNOW.
- Yandell K (2013). "Genetic Roots of the Ashkenazi Jews". The Scientist.
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (December 2002). "Genetic structure of human populations". Science. 298 (5602): 2381–5. Bibcode:2002Sci...298.2381R. doi:10.1126/science.1078311. PMID 12493913. S2CID 8127224.
- Costa MD, Pereira JB, Pala M, Fernandes V, Olivieri A, Achilli A, et al. (2013). "A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages". Nature Communications. 4: 2543. Bibcode:2013NatCo...4.2543C. doi:10.1038/ncomms3543. PMC 3806353. PMID 24104924.
- Banda Y, Kvale M, Hoffmann T, Hesselson S, Tang H, Ranatunga D, et al. (2013). "Admixture Estimation in a Founder Population". Am Soc Hum Genet. Archived from the original on 2019-08-11. Retrieved 2016-10-31.
- Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST (September 2010). "Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population". Proceedings of the National Academy of Sciences of the United States of America. 107 (37): 16222–7. Bibcode:2010PNAS..10716222B. doi:10.1073/pnas.1004381107. PMC 2941333. PMID 20798349.
- Atzmon G, Hao L, Pe'er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, Ostrer H (June 2010). "Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry". American Journal of Human Genetics. 86 (6): 850–9. doi:10.1016/j.ajhg.2010.04.015. PMC 3032072. PMID 20560205.
- "Genes Set Jews Apart, Study Finds". American Scientist. Retrieved 8 November 2013.
- "Study finds genetic links among Jewish people". EurekAlert! (Press release). 3 June 2010.
- Pala M, Achilli A, Olivieri A, Hooshiar Kashani B, Perego UA, Sanna D, et al. (June 2009). "Mitochondrial haplogroup U5b3: a distant echo of the epipaleolithic in Italy and the legacy of the early Sardinians" (PDF). American Journal of Human Genetics. 84 (6): 814–21. doi:10.1016/j.ajhg.2009.05.004. PMC 2694970. PMID 19500771. Archived from the original (PDF) on 2018-07-22. Retrieved 2010-07-15.
- Leonardi, Michela; Sandionigi, Anna; Conzato, Annalisa; Vai, Stefania; Lari, Martina (2018). "The female ancestor's tale: Long‐term matrilineal continuity in a nonisolated region of Tuscany". American Journal of Physical Anthropology. New York City: John Wiley & Sons (published September 6, 2018). 167 (3): 497–506. doi:10.1002/ajpa.23679. PMID 30187463. S2CID 52161000.
- Kovacevic L, Tambets K, Ilumäe AM, Kushniarevich A, Yunusbayev B, Solnik A, et al. (2014). "Standing at the gateway to Europe--the genetic structure of Western balkan populations based on autosomal and haploid markers". PLOS ONE. 9 (8): e105090. Bibcode:2014PLoSO...9j5090K. doi:10.1371/journal.pone.0105090. PMC 4141785. PMID 25148043.
- Wade N (13 August 2008). "Genetic Map of Europe". New York Times. Retrieved 25 February 2014.
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (February 2008). "Worldwide human relationships inferred from genome-wide patterns of variation" (PDF). Science. New York, N.Y. 319 (5866): 1100–4. Bibcode:2008Sci...319.1100L. doi:10.1126/science.1153717. PMID 18292342. S2CID 53541133. Archived from the original (PDF) on 2009-03-27.
- López Herráez D, Bauchet M, Tang K, Theunert C, Pugach I, Li J, et al. (November 2009). "Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs". PLOS ONE. 4 (11): e7888. Bibcode:2009PLoSO...4.7888L. doi:10.1371/journal.pone.0007888. PMC 2775638. PMID 19924308.
- Moorjani P, Patterson N, Hirschhorn JN, Keinan A, Hao L, Atzmon G, Burns E, Ostrer H, Price AL, Reich D (April 2011). "The history of African gene flow into Southern Europeans, Levantines, and Jews". PLOS Genetics. 7 (4): e1001373. doi:10.1371/journal.pgen.1001373. PMC 3080861. PMID 21533020.
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, et al. (February 2012). "New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing". Nature Communications. 3: 698. Bibcode:2012NatCo...3..698K. doi:10.1038/ncomms1701. PMID 22426219.
- Raveane, Alessandro; Molinaro, Ludovica; Aneli, Serena; Capodiferro, Marco Rosario; Ongaro, Linda; Migliore, Nicola Rambaldi; Soffiati, Sara; Scarano, Teodoro; Torroni, Antonio; Achilli, Alessandro; Ventura, Mario; Pagani, Luca; Capelli, Cristian; Olivieri, Anna; Bertolini, Francesco (2022-03-01). "Assessing temporal and geographic contacts across the Adriatic Sea through the analysis of genome-wide data from Southern Italy": 2022.02.26.482072. doi:10.1101/2022.02.26.482072. S2CID 247231413.
{{cite journal}}: Cite journal requires|journal=(help)
Further reading
- Saupe, Tina et al. "Ancient genomes reveal structural shifts after the arrival of Steppe-related ancestry in the Italian Peninsula". In: Current Biology Volume 31, Issue 12, 21 June 2021, Pages 2576–2591.e12. DOI: https://doi.org/10.1016/j.cub.2021.04.022