Collective cell migration
Collective cell migration describes the movements of group of cells and the emergence of collective behavior from cell-environment interactions and cell-cell communication. Collective cell migration is an essential process in the lives of multicellular organisms, e.g. embryonic development, wound healing and cancer spreading (metastasis).[1] Cells can migrate as a cohesive group (e.g. epithelial cells) or have transient cell-cell adhesion sites (e.g. mesenchymal cells).[2] They can also migrate in different modes like sheets, strands, tubes, and clusters.[3] While single-cell migration has been extensively studied, collective cell migration is a relatively new field with applications in preventing birth defects or dysfunction of embryos. It may improve cancer treatment by enabling doctors to prevent tumors from spreading and forming new tumors.
Cell-environment interactions
| Part of a series on |
| Microbial and microbot movement |
|---|
.png.webp) |
| Microswimmers |
| Molecular motors |
|
The environment of the migrating cell can affect its speed, persistence and direction of migration by stimulating it. The extracellular matrix (ECM) provides not only the structural and biochemical support, but also plays a major role in regulating cell behavior. Different ECM proteins (such as collagen, elastin, fibronectin, laminin, and others) allow cells to adhere and migrate, while forming focal adhesions in the front and disassembling them in the back. Using these adhesion sites, cells also sense the mechanical properties of the ECM. Cells can be guided by a gradient of those proteins (haptotaxis) or a gradient of soluble substrates in the liquid phase surrounding the cell (chemotaxis). Cells sense the substrate through their receptors and migrate toward the concentration (or the opposite direction). Another form of stimulation can be rigidity gradients of the ECM (durotaxis).
Confinement
Collective cell migration is enhanced by geometrical confinement of an extracellular matrix molecule (e.g. the proteoglycan versican in neural crest cells), that acts as a barrier, to promote the emergence of organized migration in separated streams. Confinement is also observed in vivo, where the optimal width is a function of the number of migrating cells in different streams of different species.[4]
Cell-cell communication
Migrating isolated cell responds to cues in its environment and changes its behavior accordingly. As cell-cell communication does not play a major role in this case, similar trajectories are observed in different isolated cells. However, when the cell migrates as part of the collective, it not only responds to its environment but also interacts with other cells through soluble substrates and physical contact. These cell-cell communication mechanisms are the main reasons for the difference between efficient migration of the collective and random walk movements of the isolated cell. Cell-cell communication mechanisms are widely studied experimentally (in vivo and in vitro),[5] and computationally (in silico).[6]
Co-attraction
Co-attraction between collectively migrating cells is the process by which cells of the same type secrete chemo-attractant (e.g. C3a in neural crest cells), that stimulates other cells in the group that have the receptors to that chemo-attractant. Cells sense the secreted substrate and respond to the stimulation by moving towards each other's and maintain high cell density.[7][8]
Contact inhibition of locomotion
Contact inhibition of locomotion (CIL) is a process in which the cell changes its direction of movement after colliding into another cell. Those cells could be of the same cell type or different types. The contacts (cell-junctions) are created by transmembrane glycoproteins named cadherins (E-cadherin, N-cadherin or cadherin 11) and other proteins. After cell-cell contact, the protrusions of cells in the contact direction are inhibited. In the CIL process, cells migrate away from each other by repolarizing in the new direction, so that new protrusions are formed in the front while contractions pull the back from contact.
- Contact inhibition of proliferation (CIP) is the inhibition of cell division with increasing percent of confluency. CIP and CIL are two different processes, which are sometimes mistakenly interrelated.[9]
Examples of studied systems
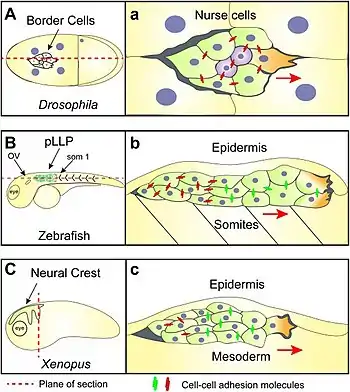
Red arrows show the direction of migration for each tissue
Collective cell migration is studied over many model species.
Border cells in flies (Drosophila melanogaster): the border cells migrate during the differentiation of egg cells to be ready for fertilization.[11]
The lateral line in zebrafish: collective cell migration from head to tails is essential to the development of the sensory system of the fish. The sensors of the lateral line measure the flow over the body-surface of the fish.[12]
Wound healing: collective cell migration is an essential part in this healing process, wound area is closed by the migrating cells.[13][14] Wound healing is commonly studied in vitro using cell lines such as Madin-Darby Canine Kidney cells.
Neural crest cells in mice,[15] Leghorn chicks,[16] amphibians (Xenopus laevis),[17] and fish[18] (zebrafish): collective migration of neural crest cells occurs during embryo development of vertebrates. They migrate long distances from the head (neural tube) to give rise to different tissues.[19]
Spreading of cancer (metastasis): common complication of cancer involve formation of new tumors (secondary tumors), as a result of migration of cancer cells from the primary tumor. Similar to collective cell migration in development and wound healing, cancer cells also undergo epithelial to mesenchymal transition (EMT), that reduces cell-cell adhesions and allows cancer spreading.[20]
The diagram on the right shows:
- A: Border cell migration in a Drosophila embryo. (a) shows border cells migrating in a confined space surrounded by gigantic nurse cells.[10]
- B: The initial position of zebrafish posterior lateral line primordia (pLLP) cells. (b) is a sagittal section showing how the these cells migrate in a confined space between the somatic mesoderm and epidermis.[10]
- C: The cephalic neural crest of the clawed frog Xenopus migrating in well-defined streams from dorsal to ventral and anterior. (c) is a transverse section across the head of a Xenopus embryo showing the high degree of confinement experienced by the neural crest while migrating sandwiched between the epidermis and underlying head mesoderm.[10]
Mathematical models
There are several mathematical models that describe collective cell motion. Typically, a Newtonian equation of motion for a system of cells is solved.[21] Several forces act on each individual cell, examples are friction (between environment and other cells), chemotaxis and self-propulsion. The latter implies that cells are active matter far from thermal equilibrium that are able to generate force due to myosin-actin contractile motion. An overview over physical description of collective cell migration[22] explains that the following types of models can be used:
- Lattice models (e.g. BIO-LGCA models)
- Models similar to Dissipative particle dynamics that solve Newton's equation of motion with dissipative and random forces
- Models where cells depict Voronoi regions and an effective potential (based on Voronoi graphs) for the tissue is used
- Continuum models, e.g. with the use of a phase field
- Kinetic theories similar to the Boltzmann equation[23]
These mathematical models give some insight in complex phenomena like cancer, wound healing[24] and ectoplasms.


Spectrum of collective cell migration
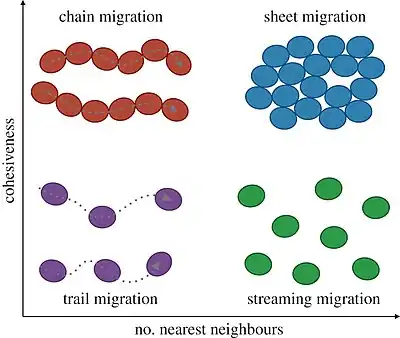
In the diagram immediately below, different morphologies of collective cell migration are characterized by their cohesiveness during migration (inversely related to density), as well as the number of nearest neighbours with which a cell interacts while moving (i.e. the topological arrangement of individual cells in the population). Cells (ellipses) can migrate in linear chains (top left), with persistent contact to cells either side of them, or along trails formed by preceding cells (bottom left). In migrating sheets, cells may maintain most of their nearest neighbours over time (top right), whereas in streaming migration cell–cell contacts occur at longer range and with potentially frequent neighbour rearrangement (bottom right). These concepts easily extend to three-dimensional migration, in which case the place of migrating sheets can be taken by moving clusters or spheroids.[25]
See also
References
- Friedl, P; Hegerfeldt, Y; Tusch, M (2004). "Collective cell migration in morphogenesis and cancer". The International Journal of Developmental Biology. 48 (5–6): 441–9. doi:10.1387/ijdb.041821pf. PMID 15349818.
- Weijer, CJ (15 September 2009). "Collective cell migration in development". Journal of Cell Science. 122 (Pt 18): 3215–23. doi:10.1242/jcs.036517. PMID 19726631.

- Friedl, P (February 2004). "Prespecification and plasticity: shifting mechanisms of cell migration". Current Opinion in Cell Biology. 16 (1): 14–23. doi:10.1016/j.ceb.2003.11.001. PMID 15037300.
- Szabó, András; Melchionda, Manuela; Nastasi, Giancarlo; Woods, Mae L.; Campo, Salvatore; Perris, Roberto; Mayor, Roberto (6 June 2016). "In vivo confinement promotes collective migration of neural crest cells". Journal of Cell Biology. 213 (5): 543–555. doi:10.1083/jcb.201602083. PMC 4896058. PMID 27241911.
- Mayor, R; Etienne-Manneville, S (February 2016). "The front and rear of collective cell migration" (PDF). Nature Reviews. Molecular Cell Biology. 17 (2): 97–109. doi:10.1038/nrm.2015.14. PMID 26726037. S2CID 27261044.
- Szabó, A; Mayor, R (October 2016). "Modelling collective cell migration of neural crest". Current Opinion in Cell Biology. 42: 22–28. doi:10.1016/j.ceb.2016.03.023. PMC 5017515. PMID 27085004.
- Woods, ML; Carmona-Fontaine, C; Barnes, CP; Couzin, ID; Mayor, R; Page, KM (2014). "Directional collective cell migration emerges as a property of cell interactions". PLoS ONE. 9 (9): e104969. Bibcode:2014PLoSO...9j4969W. doi:10.1371/journal.pone.0104969. PMC 4152153. PMID 25181349.
- Carmona-Fontaine, C; Theveneau, E; Tzekou, A; Tada, M; Woods, M; Page, KM; Parsons, M; Lambris, JD; Mayor, R (13 December 2011). "Complement fragment C3a controls mutual cell attraction during collective cell migration". Developmental Cell. 21 (6): 1026–37. doi:10.1016/j.devcel.2011.10.012. PMC 3272547. PMID 22118769.
- Stoker, MG; Rubin, H (8 July 1967). "Density dependent inhibition of cell growth in culture". Nature. 215 (5097): 171–2. Bibcode:1967Natur.215..171S. doi:10.1038/215171a0. PMID 6049107. S2CID 4150783.
- Barriga, Elias H.; Mayor, Roberto (2019). "Adjustable viscoelasticity allows for efficient collective cell migration". Seminars in Cell & Developmental Biology. 93: 55–68. doi:10.1016/j.semcdb.2018.05.027. PMC 6854469. PMID 29859995.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Bianco, A; Poukkula, M; Cliffe, A; Mathieu, J; Luque, CM; Fulga, TA; Rørth, P (19 July 2007). "Two distinct modes of guidance signalling during collective migration of border cells". Nature. 448 (7151): 362–5. Bibcode:2007Natur.448..362B. doi:10.1038/nature05965. PMID 17637670. S2CID 4369682.
- Dalle Nogare, D; Somers, K; Rao, S; Matsuda, M; Reichman-Fried, M; Raz, E; Chitnis, AB (August 2014). "Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium". Development. 141 (16): 3188–96. doi:10.1242/dev.106690. PMC 4197546. PMID 25063456.
- Grada A (February 2017). "Analysis of Collective Cell Migration Using the Wound Healing Assay". J Invest Dermatol. 137 (2): e11–e16. doi:10.1016/j.jid.2016.11.020. PMID 28110712.
- Trepat, Xavier; Wasserman, Michael R.; Angelini, Thomas E.; Millet, Emil; Weitz, David A.; Butler, James P.; Fredberg, Jeffrey J. (3 May 2009). "Physical forces during collective cell migration". Nature Physics. 5 (6): 426–430. Bibcode:2009NatPh...5..426T. doi:10.1038/nphys1269.
- Trainor, PA (December 2005). "Specification of neural crest cell formation and migration in mouse embryos". Seminars in Cell & Developmental Biology. 16 (6): 683–93. doi:10.1016/j.semcdb.2005.06.007. PMID 16043371.
- Johnston, MC (October 1966). "A radioautographic study of the migration and fate of cranial neural crest cells in the chick embryo". The Anatomical Record. 156 (2): 143–55. doi:10.1002/ar.1091560204. PMID 5969670. S2CID 11251193.
- Sadaghiani, B; Thiébaud, CH (November 1987). "Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy". Developmental Biology. 124 (1): 91–110. doi:10.1016/0012-1606(87)90463-5. PMID 3666314.
- Smith, M.; Hickman, A.; Amanze, D.; Lumsden, A.; Thorogood, P. (23 May 1994). "Trunk neural crest origin of caudal fin mesenchyme in the zebrafish Brachydanio rerio". Proceedings of the Royal Society B: Biological Sciences. 256 (1346): 137–145. Bibcode:1994RSPSB.256..137S. doi:10.1098/rspb.1994.0061. JSTOR 50346. S2CID 86494317.
- Le Douarin, Nicole, and Chaya Kalcheim. The neural crest. No. 36. Cambridge University Press, 1999.
- Thiery, JP; Acloque, H; Huang, RY; Nieto, MA (25 November 2009). "Epithelial-mesenchymal transitions in development and disease". Cell. 139 (5): 871–90. doi:10.1016/j.cell.2009.11.007. PMID 19945376.

- Akiyama, Masakazu; Sushida, Takamichi; Ishida, Sumire; Haga, Hisashi (2017). "Mathematical model of collective cell migrations based on cell polarity". Development, Growth & Differentiation. 59 (5): 471–490. doi:10.1111/dgd.12381. ISSN 1440-169X. PMID 28714585.
- Alert, Ricard; Trepat, Xavier (2020-03-10). "Physical Models of Collective Cell Migration". Annual Review of Condensed Matter Physics. 11 (1): 77–101. arXiv:1905.07675. Bibcode:2020ARCMP..11...77A. doi:10.1146/annurev-conmatphys-031218-013516. ISSN 1947-5454. S2CID 159041328.
- Chauviere, A.; Hillen, T.; Preziosi, L. (2007). "Modeling cell movement in anisotropic and heterogeneous network tissues". Networks & Heterogeneous Media. 2 (2): 333–357. doi:10.3934/nhm.2007.2.333. ISSN 1556-181X.
- Arciero, Julia C.; Mi, Qi; Branca, Maria F.; Hackam, David J.; Swigon, David (2011-02-02). "Continuum Model of Collective Cell Migration in Wound Healing and Colony Expansion". Biophysical Journal. 100 (3): 535–543. Bibcode:2011BpJ...100..535A. doi:10.1016/j.bpj.2010.11.083. ISSN 0006-3495. PMC 3030184. PMID 21281567.
- Schumacher, Linus J.; Kulesa, Paul M.; McLennan, Rebecca; Baker, Ruth E.; Maini, Philip K. (2016). "Multidisciplinary approaches to understanding collective cell migration in developmental biology". Open Biology. 6 (6): 160056. doi:10.1098/rsob.160056. PMC 4929938. PMID 27278647.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.