BIO-LGCA
In computational and mathematical biology, a biological lattice-gas cellular automaton (BIO-LGCA) is a discrete model for moving and interacting biological agents,[1] a type of cellular automaton. The BIO-LGCA is based on the lattice-gas cellular automaton (LGCA) model used in fluid dynamics. A BIO-LGCA model describes cells and other motile biological agents as point particles moving on a discrete lattice, thereby interacting with nearby particles. Contrary to classic cellular automaton models, particles in BIO-LGCA are defined by their position and velocity. This allows to model and analyze active fluids and collective migration mediated primarily through changes in momentum, rather than density. BIO-LGCA applications include cancer invasion[2] and cancer progression.[3]
Model definition
As are all cellular automaton models, a BIO-LGCA model is defined by a lattice , a state space , a neighborhood , and a rule .[4]
- The lattice () defines the set of all possible particle positions. Particles are restricted to occupy only certain positions, typically resulting from a regular and periodic tesselation of space. Mathematically, is a discrete subset of -dimensional space.
- The state space () describes the possible states of particles within every lattice site . In BIO-LGCA, multiple particles with different velocities may occupy a single lattice site, as opposed to classic cellular automaton models, where typically only a single cell can reside in every lattice node simultaneously. This makes the state space slightly more complex than that of classic cellular automaton models (see below).
- The neighborhood () indicates the subset of lattice sites which determines the dynamics of a given site in the lattice. Particles only interact with other particles within their neighborhood. Boundary conditions must be chosen for neighborhoods of lattice sites at the boundary of finite lattices. Neighborhoods and boundary conditions are identically defined as those for regular cellular automata (see Cellular automaton).
- The rule () dictates how particles move, proliferate, or die with time. As every cellular automaton, BIO-LGCA evolves in discrete time steps. In order to simulate the system dynamics, the rule is synchronously applied to every lattice site at every time step. Rule application changes the original state of a lattice site to a new state. The rule depends on the states of lattice sites in the interaction neighborhood of the lattice site to be updated. In BIO-LGCA, the rule is divided into two steps, a probabilistic interaction step followed by a deterministic transport step. The interaction step simulates reorientation, birth, and death processes, and is defined specifically for the modeled process. The transport step translocates particles to neighboring lattice nodes in the direction of their velocities. See below for details.
State space
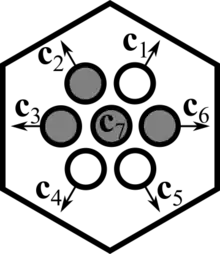
For modeling particle velocities explicitly, lattice sites are assumed to have a specific substructure. Each lattice site is connected to its neighboring lattice sites through vectors called "velocity channels", , , where the number of velocity channels is equal to the number of nearest neighbors, and thus depends on the lattice geometry ( for a one-dimensional lattice, for a two-dimensional hexagonal lattice, and so on). In two dimensions, velocity channels are defined as . Additionally, an arbitrary number of so-called "rest channels" may be defined, such that , . A channel is said to be occupied if there is a particle in the lattice site with a velocity equal to the velocity channel. The occupation of channel is indicated by the occupation number . Typically, particles are assumed to obey an exclusion principle, such that no more than one particle may occupy a single velocity channel at a lattice site simultaneously. In this case, occupation numbers are Boolean variables, i.e. , and thus, every site has a maximum carrying capacity . Since the collection of all channel occupation numbers defines the number of particles and their velocities in each lattice site, the vector describes the state of a lattice site, and the state space is given by .
Rule and model dynamics
The states of every site in the lattice are updated synchronously in discrete time steps to simulate the model dynamics. The rule is divided into two steps. The probabilistic interaction step simulates particle interaction, while the deterministic transport step simulates particle movement.
Interaction step
Depending on the specific application, the interaction step may be composed of reaction and/or reorientation operators.
The reaction operator replaces the state of a node with a new state following a transition probability , which depends on the state of the neighboring lattice sites to simulate the influence of neighboring particles on the reactive process. The reaction operator does not conserve particle number, thus allowing to simulate birth and death of individuals. The reaction operator's transition probability is usually defined ad hoc form phenomenological observations.
The reorientation operator also replaces a state with a new state with probability . However, this operator conserves particle number and therefore only models changes in particle velocity by redistributing particles among velocity channels. The transition probability for this operator can be determined from statistical observations (by using the maximum caliber principle) or from known single-particle dynamics (using the discretized, steady-state angular probability distribution given by the Fokker-Planck equation associated to a Langevin equation describing the reorientation dynamics),[5][6] and typically takes the form
where is a normalization constant (also known as the partition function), is an energy-like function which particles will likely minimize when changing their direction of motion, is a free parameter inversely proportional to the randomness of particle reorientation (analogous to the inverse temperature in thermodynamics), and is a Kronecker delta which ensures that particle number before and after reorientation is unchanged.
The state resulting form applying the reaction and reorientation operator is known as the post-interaction configuration and denoted by .
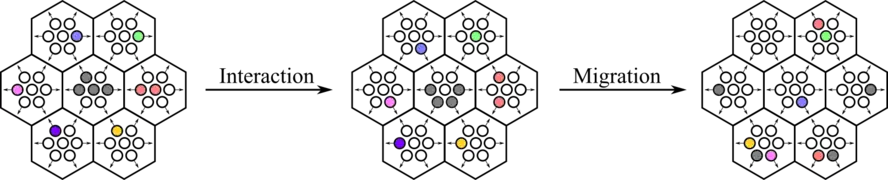
Transport step
After the interaction step, the deterministic transport step is applied synchronously to all lattice sites. The transport step simulates the movement of agents according to their velocity, due to the self-propulsion of living organisms.
During this step, the occupation numbers of post-interaction states will be defined as the new occupation states of the same channel of the neighboring lattice site in the direction of the velocity channel, i.e. .
A new time step begins when both interaction and transport steps have occurred. Therefore, the dynamics of the BIO-LGCA can be summarized as the stochastic finite-difference microdynamical equation
Example interaction dynamics
The transition probability for the reaction and/or reorientation operator must be defined to appropriately simulate the modeled system. Some elementary interactions and the corresponding transition probabilities are listed below.
Random walk
In the absence of any external or internal stimuli, cells may move randomly without any directional preference. In this case, the reorientation operator may be defined through a transition probability
where . Such transition probability allows any post-reorientation configuration with the same number of particles as the pre-reorientation configuration , to be picked uniformly.
Simple birth and death process
If organisms reproduce and die independently of other individuals (with the exception of the finite carrying capacity), then a simple birth/death process can be simulated[3] with a transition probability given by
where , are constant birth and death probabilities, respectively, is the Kronecker delta which ensures only one birth/death event happens every time step, and is the Heaviside function, which makes sure particle numbers are positive and bounded by the carrying capacity .
Adhesive interactions
Cells may adhere to one another by cadherin molecules on the cell surface. Cadherin interactions allow cells to form aggregates. The formation of cell aggregates via adhesive biomolecules can be modeled[7] by a reorientation operator with transition probabilities defined as
where is a vector pointing in the direction of maximum cell density, defined as , where is the configuration of the lattice site within the neighborhood , and is the momentum of the post-reorientation configuration, defined as . This transition probability favors post-reorientation configurations with cells moving towards the cell density gradient.
Mathematical analysis
Since an exact treatment of a stochastic agent-based model quickly becomes unfeasible due to high-order correlations between all agents,[8] the general method of analyzing a BIO-LGCA model is to cast it into an approximate, deterministic finite difference equation (FDE) describing the mean dynamics of the population, then performing the mathematical analysis of this approximate model, and comparing the results to the original BIO-LGCA model.
First, the expected value of the microdynamical equation is obtained
where denotes the expected value, and is the expected value of the -th channel occupation number of the lattice site at at time step . However, the term on the right, is highly nonlinear on the occupation numbers of both the lattice site and the lattice sites within the interaction neighborhood , due to the form of the transition probability and the statistics of particle placement within velocity channels (for example, arising from an exclusion principle imposed on channel occupations). This non-linearity would result in high-order correlations and moments among all channel occupations involved. Instead, a mean-field approximation is usually assumed, wherein all correlations and high order moments are neglected, such that direct particle-particle interactions are substituted by interactions with the respective expected values. In other words, if are random variables, and is a function, then under this approximation. Thus, we can simplify the equation to
where is a nonlinear function of the expected lattice site configuration and the expected neighborhood configuration dependent on the transition probabilities and in-node particle statistics. From this nonlinear FDE, one may identify several homogeneous steady states, or constants independent of and which are solutions to the FDE. To study the stability conditions of these steady states and the pattern formation potential of the model, a linear stability analysis can be performed. To do so, the nonlinear FDE is linearized as
where denotes the homogeneous steady state , and a von Neumann neighborhood was assumed. In order to cast it into a more familiar finite difference equation with temporal increments only, a discrete Fourier transform can be applied on both sides of the equation. After applying the shift theorem and isolating the term with a temporal increment on the left, one obtains the lattice-Boltzmann equation[4]
where is the imaginary unit, is the size of the lattice along one dimension, is the Fourier wave number, and denotes the discrete Fourier transform. In matrix notation, this equation is simplified to , where the matrix is called the Boltzmann propagator and is defined as
The eigenvalues of the Boltzmann propagator dictate the stability properties of the steady state:[4]
- If , where denotes the modulus, then perturbations with wave number grow with time. If , and , then perturbations with wave number will dominate and patterns with a clear wavelength will be observed. Otherwise, the steady state is stable and any perturbations will decay.
- If , where denotes the argument, then perturbations are transported and non-stationary population behaviors are observed. Otherwise, the population will appear static at the macroscopic level.
Applications
Constructing a BIO-LGCA for the study of biological phenomena mainly involves defining appropriate transition probabilities for the interaction operator, though precise definitions of the state space (to consider several cellular phenotypes, for example), boundary conditions (for modeling phenomena in confined conditions), neighborhood (to match experimental interaction ranges quantitatively), and carrying capacity (to simulate crowding effects for given cell sizes) may be important for specific applications. While the distribution of the reorientation operator can be obtained through the aforementioned statistical and biophysical methods, the distribution of the reaction operators can be estimated from the statistics of in vitro experiments, for example.[9]
BIO-LGCA models have been used to study several cellular, biophysical and medical phenomena. Some examples include:
- Angiogenesis:[10] an in vitro experiment with endothelial cells and BIO-LGCA simulation observables were compared to determine the processes involved during angiogenesis and their weight. They found that adhesion, alignment, contact guidance, and ECM remodeling are all involved in angiogenesis, while long-range interactions are not vital to the process.
- Active fluids:[11] the macroscopic physical properties of a population of particles interacting through polar alignment interactions were investigated using a BIO-LGCA model. It was found that increasing initial particle density and interaction strength result in a second order phase transition from a homogeneous, disordered state to an ordered, patterned, moving state.
- Epidemiology:[12] a spatial SIR BIO-LGCA model was used to study the effect of different vaccination strategies, and the effect of approximating a spatial epidemic with a non-spatial model. They found that barrier-type vaccination strategies are much more effective than spatially uniform vaccination strategies. Furthermore, they found that non-spatial models greatly overestimate the rate of infection.
- Cell jamming:[13] in vitro and Bio-LGCA models were used for studying metastatic behavior in breast cancer. The BIO-LGCA model revealed that metastasis may exhibit different behaviors, such as random gas-like, jammed solid-like, and correlated fluid-like states, depending on the adhesivity level among cells, ECM density, and cell-ECM interactions.
References
- Deutsch, Andreas; Nava-Sedeño, Josué Manik; Syga, Simon; Hatzikirou, Haralampos (2021-06-15). "BIO-LGCA: A cellular automaton modelling class for analysing collective cell migration". PLOS Computational Biology. 17 (6): e1009066. Bibcode:2021PLSCB..17E9066D. doi:10.1371/journal.pcbi.1009066. ISSN 1553-7358. PMC 8232544. PMID 34129639.
- Reher, David; Klink, Barbara; Deutsch, Andreas; Voss-Böhme, Anja (2017-08-11). "Cell adhesion heterogeneity reinforces tumour cell dissemination: novel insights from a mathematical model". Biology Direct. 12 (1): 18. doi:10.1186/s13062-017-0188-z. ISSN 1745-6150. PMC 5553611. PMID 28800767.
- Böttger, Katrin; Hatzikirou, Haralambos; Voss-Böhme, Anja; Cavalcanti-Adam, Elisabetta Ada; Herrero, Miguel A.; Deutsch, Andreas (2015-09-03). Alber, Mark S (ed.). "An Emerging Allee Effect Is Critical for Tumor Initiation and Persistence". PLOS Computational Biology. 11 (9): e1004366. Bibcode:2015PLSCB..11E4366B. doi:10.1371/journal.pcbi.1004366. ISSN 1553-7358. PMC 4559422. PMID 26335202.
- "Mathematical Modeling of Biological Pattern Formation", Cellular Automaton Modeling of Biological Pattern Formation, Modeling and Simulation in Science, Engineering and Technology, Boston, MA: Birkhäuser Boston, pp. 45–56, 2005, doi:10.1007/0-8176-4415-6_3, ISBN 978-0-8176-4281-5, retrieved 2021-05-25
- Nava-Sedeño, J. M.; Hatzikirou, H.; Peruani, F.; Deutsch, A. (2017-02-27). "Extracting cellular automaton rules from physical Langevin equation models for single and collective cell migration". Journal of Mathematical Biology. 75 (5): 1075–1100. doi:10.1007/s00285-017-1106-9. ISSN 0303-6812. PMID 28243720. S2CID 32456636.
- Nava-Sedeño, J. M.; Hatzikirou, H.; Klages, R.; Deutsch, A. (2017-12-05). "Cellular automaton models for time-correlated random walks: derivation and analysis". Scientific Reports. 7 (1): 16952. arXiv:1802.04201. Bibcode:2017NatSR...716952N. doi:10.1038/s41598-017-17317-x. ISSN 2045-2322. PMC 5717221. PMID 29209065.
- Bussemaker, Harmen J. (1996-02-01). "Analysis of a pattern-forming lattice-gas automaton: Mean-field theory and beyond". Physical Review E. 53 (2): 1644–1661. Bibcode:1996PhRvE..53.1644B. doi:10.1103/physreve.53.1644. ISSN 1063-651X. PMID 9964425.
- Ovaskainen, Otso; Somervuo, Panu; Finkelshtein, Dmitri (2020-10-28). "A general mathematical method for predicting spatio-temporal correlations emerging from agent-based models". Journal of the Royal Society Interface. 17 (171): 20200655. doi:10.1098/rsif.2020.0655. PMC 7653394. PMID 33109018.
- Dirkse, Anne; Golebiewska, Anna; Buder, Thomas; Nazarov, Petr V.; Muller, Arnaud; Poovathingal, Suresh; Brons, Nicolaas H. C.; Leite, Sonia; Sauvageot, Nicolas; Sarkisjan, Dzjemma; Seyfrid, Mathieu (2019-04-16). "Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment". Nature Communications. 10 (1): 1787. Bibcode:2019NatCo..10.1787D. doi:10.1038/s41467-019-09853-z. ISSN 2041-1723. PMC 6467886. PMID 30992437.
- Mente, Carsten; Prade, Ina; Brusch, Lutz; Breier, Georg; Deutsch, Andreas (2010-10-01). "Parameter estimation with a novel gradient-based optimization method for biological lattice-gas cellular automaton models". Journal of Mathematical Biology. 63 (1): 173–200. doi:10.1007/s00285-010-0366-4. ISSN 0303-6812. PMID 20886214. S2CID 12404555.
- Bussemaker, Harmen J.; Deutsch, Andreas; Geigant, Edith (1997-06-30). "Mean-Field Analysis of a Dynamical Phase Transition in a Cellular Automaton Model for Collective Motion". Physical Review Letters. 78 (26): 5018–5021. arXiv:physics/9706008. Bibcode:1997PhRvL..78.5018B. doi:10.1103/PhysRevLett.78.5018. ISSN 0031-9007. S2CID 45979152.
- Fuks, Henryk; Lawniczak, Anna T. (2001). "Individual-based lattice model for spatial spread of epidemics". Discrete Dynamics in Nature and Society. 6 (3): 191–200. doi:10.1155/s1026022601000206.
- Ilina, Olga; Gritsenko, Pavlo G.; Syga, Simon; Lippoldt, Jürgen; La Porta, Caterina A. M.; Chepizhko, Oleksandr; Grosser, Steffen; Vullings, Manon; Bakker, Gert-Jan; Starruß, Jörn; Bult, Peter (2020-08-24). "Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion". Nature Cell Biology. 22 (9): 1103–1115. doi:10.1038/s41556-020-0552-6. ISSN 1476-4679. PMC 7502685. PMID 32839548.
External links
- Bio-LGCA Simulator - An online simulator with elementary interactions with personalizable parameter values.
- BIO-LGCA Python Package - An open source Python package for implementing BIO-LGCA model simulations.