Vesiculovirus matrix proteins
The family of vesiculovirus matrix proteins consists of several matrix proteins of the vesicular stomatitis virus, also known as VSIV or VSV. The matrix (M) protein of the virus causes many of the cytopathic effects of VSV, including an inhibition of host gene expression and the induction of cell rounding. It has been shown that M protein also induces apoptosis in the absence of other viral components. It is thought that the activation of apoptotic pathways causes the inhibition of host gene expression and cell rounding by M protein.[1]
| Vesiculo_matrix | |||||||||
|---|---|---|---|---|---|---|---|---|---|
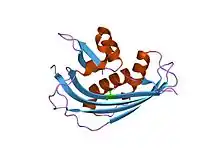 crystal structure of vesicular stomatitis virus matrix protein | |||||||||
| Identifiers | |||||||||
| Symbol | Vesiculo_matrix | ||||||||
| Pfam | PF06326 | ||||||||
| InterPro | IPR009397 | ||||||||
| SCOP2 | 1lg7 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 381 | ||||||||
| OPM protein | 1lg7 | ||||||||
| |||||||||
Function
These proteins play a major role in assembly and budding of VSIV virions.[2] Their main role is to aid virus assembly. They starts by shutting off host cell transcription by inhibiting mRNA nuclear export through direct interaction with the host RAE1-NUP98 complex. This inhibits interferon signaling and thus establishment of antiviral state in virus infected cells. In turn, this induces cell-rounding, cytoskeleton disorganization and apoptosis in infected cell. Enveloped viruses acquire their membrane by budding at a membrane of their host cell.[3]
Structure
The structure of these matrix proteins has revealed a single-globular domain with a new fold. The N-terminal part consists of a large five-stranded anti-parallel beta-sheet packed against two alpha-helices; the C-terminal part comprises a small two stranded anti-parallel beta-sheet and an alpha-helix.
References
- Kopecky SA, Lyles DS (May 2003). "The cell-rounding activity of the vesicular stomatitis virus matrix protein is due to the induction of cell death". J. Virol. 77 (9): 5524–8. doi:10.1128/jvi.77.9.5524-5528.2003. PMC 153969. PMID 12692256.
- Gaudier, Martin; Gaudin, Yves; Knossow, Marcel (2002-06-17). "Crystal structure of vesicular stomatitis virus matrix protein". The EMBO Journal. 21 (12): 2886–2892. doi:10.1093/emboj/cdf284. ISSN 0261-4189. PMC 126044. PMID 12065402.
- Solon J, Gareil O, Bassereau P, Gaudin Y (2005). "Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system". J Gen Virol. 86 (Pt 12): 3357–63. doi:10.1099/vir.0.81129-0. PMID 16298982.