Human genetic clustering
Human genetic clustering refers to patterns of relative genetic similarity among human individuals and populations, as well as the wide range of scientific and statistical methods used to study this aspect of human genetic variation.
Clustering studies are thought to be valuable for characterizing the general structure of genetic variation among human populations, to contribute to the study of ancestral origins, evolutionary history, and precision medicine. Since the mapping of the human genome, and with the availability of increasingly powerful analytic tools, cluster analyses have revealed a range of ancestral and migratory trends among human populations and individuals.[1] Human genetic clusters tend to be organized by geographic ancestry, with divisions between clusters aligning largely with geographic barriers such as oceans or mountain ranges.[2][3] Clustering studies have been applied to global populations,[4] as well as to population subsets like post-colonial North America.[5][6] Notably, the practice of defining clusters among modern human populations is largely arbitrary and variable due to the continuous nature of human genotypes; although individual genetic markers can be used to produce smaller groups, there are no models that produce completely distinct subgroups when larger numbers of genetic markers are used.[2][7][8]
Many studies of human genetic clustering have been implicated in discussions of race, ethnicity, and scientific racism, as some have controversially suggested that genetically derived clusters may be understood as proof of genetically determined races.[9][10] Although cluster analyses invariably organize humans (or groups of humans) into subgroups, debate is ongoing on how to interpret these genetic clusters with respect to race and its social and phenotypic features. And, because there is such a small fraction of genetic variation between human genotypes overall, genetic clustering approaches are highly dependent on the sampled data, genetic markers, and statistical methods applied to their construction.
Genetic clustering algorithms and methods
A wide range of methods have been developed to assess the structure of human populations with the use of genetic data. Early studies of within and between-group genetic variation used physical phenotypes and blood groups, with modern genetic studies using genetic markers such as Alu sequences, short tandem repeat polymorphisms, and single nucleotide polymorphisms (SNPs), among others.[11] Models for genetic clustering also vary by algorithms and programs used to process the data. Most sophisticated methods for determining clusters can be categorized as model-based clustering methods (such as the algorithm STRUCTURE[12]) or multidimensional summaries (typically through principal component analysis).[1][13] By processing a large number of SNPs (or other genetic marker data) in different ways, both approaches to genetic clustering tend to converge on similar patterns by identifying similarities among SNPs and/or haplotype tracts to reveal ancestral genetic similarities.[13]
Model-based clustering
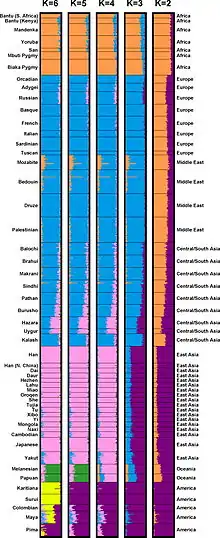
Common model-based clustering algorithms include STRUCTURE, ADMIXTURE, and HAPMIX. These algorithms operate by finding the best fit for genetic data among an arbitrary or mathematically derived number of clusters, such that differences within clusters are minimized and differences between clusters are maximized. This clustering method is also referred to as "admixture inference," as individual genomes (or individuals within populations) can be characterized by the proportions of alleles linked to each cluster.[1] In other words, algorithms like STRUCTURE generate results that assume the existence of discrete ancestral populations, operationalized through unique genetic markers, which have combined over time to form the admixed populations of the modern day.
Multidimensional summary statistics
Where model-based clustering characterizes populations using proportions of presupposed ancestral clusters, multidimensional summary statistics characterize populations on a continuous spectrum. The most common multidimensional statistical method used for genetic clustering is principal component analysis (PCA), which plots individuals by two or more axes (their "principal components") that represent aggregations of genetic markers that account for the highest variance. Clusters can then be identified by visually assessing the distribution of data; with larger samples of human genotypes, data tends to cluster in distinct groups as well as admixed positions between groups.[1][13]
Caveats and limitations
There are caveats and limitations to genetic clustering methods of any type, given the degree of admixture and relative similarity within the human population. All genetic cluster findings are biased by the sampling process used to gather data, and by the quality and quantity of that data. For example, many clustering studies use data derived from populations that are geographically distinct and far apart from one another, which may present an illusion of discrete clusters where, in reality, populations are much more blended with one another when intermediary groups are included.[1] Sample size also plays an important moderating role on cluster findings, as different sample size inputs can influence cluster assignment, and more subtle relationships between genotypes may only emerge with larger sample sizes.[1][8] In particular, the use of STRUCTURE has been widely criticized as being potentially misleading through requiring data to be sorted into a predetermined number of clusters which may or may not reflect the actual population's distribution.[8][14] The creators of STRUCTURE originally described the algorithm as an "exploratory" method to be interpreted with caution and not as a test with statistically significant power.[12][15]
Notable applications to human genetic data
Modern applications of genetic clustering methods to global-scale genetic data were first marked by studies associated with the Human Genome Diversity Project (HGDP) data.[1] These early HGDP studies, such as those by Rosenberg et al. (2002),[4][16] contributed to theories of the serial founder effect and early human migration out of Africa, and clustering methods have been notably applied to describe admixed continental populations.[5][6][17] Genetic clustering and HGDP studies have also contributed to methods for, and criticisms of, the genetic ancestry consumer testing industry.[18]
A number of landmark genetic cluster studies have been conducted on global human populations since 2002, including the following:
| Authors | Year | Title | Sample size / number of populations sampled | Sample | Markers |
|---|---|---|---|---|---|
| Rosenberg et al. | 2002 | Genetic Structure of Human Populations[19] | 1056 / 52 | Human Genome Diversity Project (HGDP-CEPH) | 377 STRs |
| Serre & Pääbo | 2004 | Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation[20] | 89 / 15 | a: HGDP | 20 STRs |
| 90 / geographically distributed individuals | b: Jorde 1997 | ||||
| Rosenberg et al. | 2005 | Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure[21] | 1056 / 52 | Human Genome Diversity Project (HGDP-CEPH) | 783 STRs + 210 indels |
| Li et al. | 2008 | Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation[22] | 938 / 51 | Human Genome Diversity Project (HGDP-CEPH) | 650,000 SNPs |
| Tishkoff et al. | 2009 | The Genetic Structure and History of Africans and African Americans[23] | ~3400 / 185 | HGDP-CEPH plus 133 additional African populations and Indian individuals | 1327 STRs + indels |
| Xing et al. | 2010 | Toward a more uniform sampling of human genetic diversity: A survey of worldwide populations by high-density genotyping[24] | 850 / 40 | HapMap plus 296 individuals | 250,000 SNPs |
Genetic clustering and race
Clusters of individuals are often geographically structured. For example, when clustering a population of East Asians and Europeans, each group will likely form its own respective cluster based on similar allele frequencies. In this way, clusters can have a correlation with traditional concepts of race and self-identified ancestry; in some cases, such as medical questionnaires, the latter variables can be used as a proxy for genetic ancestry where genetic data is unavailable.[9][4] However, genetic variation is distributed in a complex, continuous, and overlapping manner, so this correlation is imperfect and the use of racial categories in medicine can introduce additional hazards.[9]
Some scholars have challenged the idea that race can be inferred by genetic clusters, drawing distinctions between arbitrarily assigned genetic clusters, ancestry, and race. One recurring caution against thinking of human populations in terms of clusters is the notion that genotypic variation and traits are distributed evenly between populations, along gradual clines rather than along discrete population boundaries; so although genetic similarities are usually organized geographically, their underlying populations have never been completely separated from one another. Due to migration, gene flow, and baseline homogeneity, features between groups are extensively overlapping and intermixed.[2][9] Moreover, genetic clusters do not typically match socially defined racial groups; many commonly understood races may not be sorted into the same genetic cluster, and many genetic clusters are made up of individuals who would have distinct racial identities.[7] In general, clusters may most simply be understood as products of the methods used to sample and analyze genetic data; not without meaning for understanding ancestry and genetic characteristics, but inadequate to fully explaining the concept of race, which is more often described in terms of social and cultural forces.
In the related context of personalized medicine, race is currently listed as a risk factor for a wide range of medical conditions with genetic and non-genetic causes. Questions have emerged regarding whether or not genetic clusters support the idea of race as a valid construct to apply to medical research and treatment of disease, because there are many diseases that correspond with specific genetic markers and/or with specific populations, as seen with Tay-Sachs disease or sickle cell disease.[3][25] Researchers are careful to emphasize that ancestry—revealed in part through cluster analyses—plays an important role in understanding risk of disease. But racial or ethnic identity does not perfectly align with genetic ancestry, and so race and ethnicity do not reveal enough information to make a medical diagnosis.[25] Race as a variable in medicine is more likely to reflect social factors, where ancestry information is more likely to be meaningful when considering genetic ancestry.[2][25]
References
- Novembre, John; Ramachandran, Sohini (2011-09-22). "Perspectives on Human Population Structure at the Cusp of the Sequencing Era". Annual Review of Genomics and Human Genetics. 12 (1): 245–274. doi:10.1146/annurev-genom-090810-183123. ISSN 1527-8204. PMID 21801023.
- Maglo, Koffi N.; Mersha, Tesfaye B.; Martin, Lisa J. (2016-02-17). "Population Genomics and the Statistical Values of Race: An Interdisciplinary Perspective on the Biological Classification of Human Populations and Implications for Clinical Genetic Epidemiological Research". Frontiers in Genetics. 7: 22. doi:10.3389/fgene.2016.00022. ISSN 1664-8021. PMC 4756148. PMID 26925096.
- Goodman, Alan H.; Moses, Yolanda T.; Jones, Joseph L., eds. (2012-10-29). Race. doi:10.1002/9781118233023. ISBN 9781118233023.
- Rosenberg, N. A. (2002-12-20). "Genetic Structure of Human Populations". Science. 298 (5602): 2381–2385. Bibcode:2002Sci...298.2381R. doi:10.1126/science.1078311. ISSN 0036-8075. PMID 12493913. S2CID 8127224.
- Han, Eunjung; Carbonetto, Peter; Curtis, Ross E.; Wang, Yong; Granka, Julie M.; Byrnes, Jake; Noto, Keith; Kermany, Amir R.; Myres, Natalie M.; Barber, Mathew J.; Rand, Kristin A. (2017-02-07). "Clustering of 770,000 genomes reveals post-colonial population structure of North America". Nature Communications. 8 (1): 14238. Bibcode:2017NatCo...814238H. doi:10.1038/ncomms14238. ISSN 2041-1723. PMC 5309710. PMID 28169989.
- Jordan, I. King; Rishishwar, Lavanya; Conley, Andrew B. (September 2019). "Native American admixture recapitulates population-specific migration and settlement of the continental United States". PLOS Genetics. 15 (9): e1008225. doi:10.1371/journal.pgen.1008225. ISSN 1553-7404. PMC 6756731. PMID 31545791.
- Bamshad, Michael J.; Olson, Steve E. (December 2003). "Does Race Exist?". Scientific American. 289 (6): 78–85. Bibcode:2003SciAm.289f..78B. doi:10.1038/scientificamerican1203-78. ISSN 0036-8733. PMID 14631734.
- Kalinowski, S T (2010-08-04). "The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure". Heredity. 106 (4): 625–632. doi:10.1038/hdy.2010.95. ISSN 0018-067X. PMC 3183908. PMID 20683484.
- Jorde, Lynn B; Wooding, Stephen P (2004-10-26). "Genetic variation, classification and 'race'". Nature Genetics. 36 (S11): S28–S33. doi:10.1038/ng1435. ISSN 1061-4036. PMID 15508000.
- Marks, Jonathan (27 February 2017). Is science racist?. John Wiley & Sons. ISBN 978-0-7456-8925-8. OCLC 1037867598.
- Bamshad, Michael; Wooding, Stephen; Salisbury, Benjamin A.; Stephens, J. Claiborne (August 2004). "Deconstructing the relationship between genetics and race". Nature Reviews Genetics. 5 (8): 598–609. doi:10.1038/nrg1401. ISSN 1471-0056. PMID 15266342. S2CID 12378279.
- Pritchard, Jonathan K; Stephens, Matthew; Donnelly, Peter (2000-06-01). "Inference of Population Structure Using Multilocus Genotype Data". Genetics. 155 (2): 945–959. doi:10.1093/genetics/155.2.945. ISSN 1943-2631. PMC 1461096. PMID 10835412.
- Lawson, Daniel John; Falush, Daniel (2012-09-22). "Population Identification Using Genetic Data". Annual Review of Genomics and Human Genetics. 13 (1): 337–361. doi:10.1146/annurev-genom-082410-101510. ISSN 1527-8204. PMID 22703172.
- Lawson, Daniel J.; van Dorp, Lucy; Falush, Daniel (2018-08-14). "A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots". Nature Communications. 9 (1): 3258. Bibcode:2018NatCo...9.3258L. doi:10.1038/s41467-018-05257-7. ISSN 2041-1723. PMC 6092366. PMID 30108219.
- Novembre, John (2016-10-01). "Pritchard, Stephens, and Donnelly on Population Structure". Genetics. 204 (2): 391–393. doi:10.1534/genetics.116.195164. ISSN 1943-2631. PMC 5068833. PMID 27729489.
- Rosenberg, Noah A; Mahajan, Saurabh; Ramachandran, Sohini; Zhao, Chengfeng; Pritchard, Jonathan K; Feldman, Marcus W (2005-12-09). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genetics. 1 (6): e70. doi:10.1371/journal.pgen.0010070. ISSN 1553-7404. PMC 1310579. PMID 16355252.
- Leslie, Stephen; Winney, Bruce; Hellenthal, Garrett; Davison, Dan; Boumertit, Abdelhamid; Day, Tammy; Hutnik, Katarzyna; Royrvik, Ellen C.; Cunliffe, Barry; Lawson, Daniel J.; Falush, Daniel (March 2015). "The fine-scale genetic structure of the British population". Nature. 519 (7543): 309–314. Bibcode:2015Natur.519..309.. doi:10.1038/nature14230. ISSN 1476-4687. PMC 4632200. PMID 25788095.
- Royal, Charmaine D.; Novembre, John; Fullerton, Stephanie M.; Goldstein, David B.; Long, Jeffrey C.; Bamshad, Michael J.; Clark, Andrew G. (2010-05-14). "Inferring Genetic Ancestry: Opportunities, Challenges, and Implications". American Journal of Human Genetics. 86 (5): 661–673. doi:10.1016/j.ajhg.2010.03.011. ISSN 0002-9297. PMC 2869013. PMID 20466090.
- Rosenberg, Noah A.; Pritchard, Jonathan K.; Weber, James L.; Cann, Howard M.; Kidd, Kenneth K.; Zhivotovsky, Lev A.; Feldman, Marcus W. (2002-12-20). "Genetic Structure of Human Populations". Science. 298 (5602): 2381–2385. Bibcode:2002Sci...298.2381R. doi:10.1126/science.1078311. ISSN 0036-8075. PMID 12493913. S2CID 8127224.
- Serre, David; Pääbo, Svante (September 2004). "Evidence for gradients of human genetic diversity within and among continents". Genome Research. 14 (9): 1679–1685. doi:10.1101/gr.2529604. ISSN 1088-9051. PMC 515312. PMID 15342553.
- Rosenberg, NA; Mahajan, S; Ramachandran, S; Zhao, C; Pritchard, JK; et al. (2005). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genet. 1 (6): e70. doi:10.1371/journal.pgen.0010070. PMC 1310579. PMID 16355252.
- Li, Jun Z.; Absher, Devin M.; Tang, Hua; Southwick, Audrey M.; Casto, Amanda M.; Ramachandran, Sohini; Cann, Howard M.; Barsh, Gregory S.; Feldman, Marcus; Cavalli-Sforza, Luigi L.; Myers, Richard M. (2008-02-22). "Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation". Science. 319 (5866): 1100–1104. Bibcode:2008Sci...319.1100L. doi:10.1126/science.1153717. ISSN 0036-8075. PMID 18292342. S2CID 53541133.
- Tishkoff, Sarah A; Reed, Floyd A; Friedlaender, Françoise R; Ehret, Christopher; Ranciaro, Alessia; Froment, Alain; Hirbo, Jibril B; Awomoyi, Agnes A; Bodo, Jean-Marie; Doumbo, Ogobara; Ibrahim, Muntaser; Juma, Abdalla T; Kotze, Maritha J; Lema, Godfrey; Moore, Jason H; Mortensen, Holly; Nyambo, Thomas B; Omar, Sabah A; Powell, Kweli; Pretorius, Gideon S; Smith, Michael W; Thera, Mahamadou A; Wambebe, Charles; Weber, James L; Williams, Scott M (2009-05-22). "The Genetic Structure and History of Africans and African Americans". Science. 324 (5930): 1035–1044. Bibcode:2009Sci...324.1035T. doi:10.1126/science.1172257. ISSN 0036-8075. PMC 2947357. PMID 19407144.
- Xing, Jinchuan; Watkins, W. Scott; Shlien, Adam; Walker, Erin; Huff, Chad D.; Witherspoon, David J.; Zhang, Yuhua; Simonson, Tatum S.; Weiss, Robert B.; Schiffman, Joshua D.; Malkin, David; Woodward, Scott R.; Jorde, Lynn B. (October 2010). "Toward a more uniform sampling of human genetic diversity: A survey of worldwide populations by high-density genotyping". Genomics. 96 (4): 199–210. doi:10.1016/j.ygeno.2010.07.004. ISSN 0888-7543. PMC 2945611. PMID 20643205.
- Koenig, Barbara A. Lee; Soo-Jin, Sandra; Richardson, Sarah S. (2008). Revisiting race in a genomic age. Rutgers University Press. ISBN 978-0-8135-4323-9. OCLC 468194495.