Influenza B virus
Influenza B virus is the only species in the genus Betainfluenzavirus in the virus family Orthomyxoviridae.
| Influenza B virus | |
|---|---|
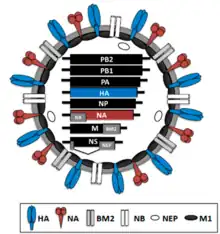 | |
| Virion structure of influenza B virus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Orthomyxoviridae |
| Genus: | Betainfluenzavirus |
| Species: | Influenza B virus |
| Synonyms | |
| |
Influenza B virus is known only to infect humans, pigs and seals.[4][5] This limited host range is apparently responsible for the lack of associated influenza pandemics in contrast with those caused by the morphologically similar influenza A virus as both mutate by both antigenic drift and reassortment.[6][7][8] There are two known circulating lineages of Influenza B virus based on the antigenic properties of the surface glycoprotein hemagglutinin. The lineages are termed B/Yamagata/16/88-like and B/Victoria/2/87-like viruses.[9] The quadrivalent influenza vaccine licensed by the CDC is currently designed to protect against both co-circulating lineages and has been shown to have greater effectiveness in prevention of influenza caused by Influenza B virus than the previous trivalent vaccine.[10]
Further diminishing the impact of this virus, "in humans, influenza B viruses evolve slower than A viruses and faster than C viruses".[11] Influenzavirus B mutates at a rate 2 to 3 times slower than type A.[12] Nevertheless, it is accepted that Influenza B virus could cause significant morbidity and mortality worldwide, and significantly impacts adolescents and schoolchildren.[13]
The B/Yamagata lineage might have become extinct in 2020/2021 due to COVID-19 pandemic measures,[14][15] and there have been no naturally occurring cases confirmed since March 2020.[16][17] In 2023, the World Health Organization concluded that protection against the Yamagata lineage was no longer necessary in the seasonal flu vaccine, reducing the number of lineages targeted by the vaccine from four to three.[16][17]
Morphology
The Influenza B virus capsid is enveloped while its virion consists of an envelope, a matrix protein, a nucleoprotein complex, a nucleocapsid, and a polymerase complex. It is sometimes spherical and sometimes filamentous. Its 500 or so surface projections are made of hemagglutinin and neuraminidase.[18]
Genome structure and genetics
The Influenza B virus genome is 14,548 nucleotides long and consists of eight segments of linear negative-sense, single-stranded RNA. The multipartite genome is encapsidated, each segment in a separate nucleocapsid, and the nucleocapsids are surrounded by one envelope.[18]
The subtypes of influenza A virus are estimated to have diverged 2,000 years ago. Influenza viruses A and B are estimated to have diverged from a single ancestor around 4,000 years ago, while the ancestor of influenza viruses A and B and the ancestor of influenza virus C are estimated to have diverged from a common ancestor around 8,000 years ago.[19] Metatranscriptomics studies have also identified closely related "Influenza B-like" viruses such as the Wuhan spiny eel influenza virus[20] and also "Influenza-B like" viruses in a number of vertebrate species such as salamanders and fish.[21]
Vaccine
In 1936, Thomas Francis Jr. discovered the ferret influenza B virus. Also in 1936, Macfarlane Burnet made the discovery that influenza virus may be cultured in hen embryonated eggs.[22] This prompted research into the properties of the virus and the creation and application of inactivated vaccines in the late 1930s and early 1940s. Inactivated vaccines' usefulness as a preventative measure was proven in the 1950s. Later, 2003 saw the approval of the first live, attenuated influenza vaccine.[22] Looking into influenza B specifically, Thomas Francis Jr. isolated influenza B virus in 1936. However, it was not until 1940 when influenza B viruses were discovered.[23]
In 1942, a new bivalent vaccine was developed that protected against both the H1N1 strain of influenza A and the newly discovered influenza B virus.[24] In today’s current world, even while some technology has advanced and flu vaccines now cover both strains of influenza A and B, the science is still based on findings from almost a century ago.[25] The viruses included in flu vaccines are changed each year to match the strains of flu that are most likely to make people sick that year since flu viruses can develop swiftly and new mutations have appeared each year, like H1N1.[25]
Even though there are two different lineages of influenza B viruses that circulate during most seasons, flu vaccinations were long meant to protect against three different flu viruses: the influenza A(H1N1), influenza A(H3N2), and one influenza B virus.[26] The second lineage of the B virus was since added to provide greater defense against circulating flu viruses.[26] Two influenza A viruses and two influenza B viruses are among the four flu viruses that a quadrivalent vaccine is intended to protect against. All flu vaccines in the United States today are quadrivalent.[26] The four main types of Type A and B influenza viruses that are most likely to spread and make people sick during the upcoming flu season are the targets of seasonal influenza (flu) vaccines.[26] All of the available flu vaccinations in the United States offer protection against the influenza A(H1), A(H3), B/Yamagata, and B/Victoria lineage viruses. Each of these four vaccine virus components is chosen based on the following criteria: which flu viruses are infecting people ahead of the upcoming flu season, how widely they are spreading, how well the vaccines from the previous flu season may protect against those flu viruses, and the vaccine viruses' capacity to offer cross-protection.[26]
For the 2022-2023 flu season, there are three flu vaccines that are preferentially recommended for people 65 years and older; various influenza (flu) vaccinations are authorized for use in people of various age groups.[26] On March 3, 2022, the FDA's Vaccines and Related Biological Products Advisory Committee (VRBPAC) convened in Silver Spring, Maryland, to choose the influenza viruses that will make up the influenza vaccine for the 2022–2023 influenza season in the United States. The committee proposed using A(H1N1)pdm09, A(H3N2), and B/Austria/1359417/2021-like viruses for trivalent influenza vaccines to be utilized in the U.S. for the 2022–2023 influenza season.[27]
Discovery and development
In 1940, an acute respiratory illness outbreak in Northern America led to the discovery of influenza B virus (IBV), which was later discovered to not have any antigenic cross-reactivity with influenza A virus (IAV). Based on calculations of the rate of amino acid substitutions in HA proteins, it was estimated that IBV and IAV diverged from one another around 4000 years ago.[28] However, the mechanisms of replication and transcription, as well as the functionality of the majority of viral proteins, appear to be largely conserved, with some unusual differences.[29] Although IBV has occasionally been found in seals and pigs, its primary host species is the human.[30] IBVs can also spread epidemics throughout the world, but they receive less attention than IAVs do due to their less prevalent nature, both in infecting hosts and in the symptoms that result from infection. IBVs used to be unclassified, but since the 1980s, they have been divided into the B/Yamagata and B/Victoria lineages.[31] IBVs have further divisions known as clades and sub-clades, just like IAVs do.[31]
Hemagglutinin (HA) and neuraminidase (NA) are two virus surface antigens that are constantly changing.[22] Antigenic drift or antigenic shift are two possible influenza viral changes. Small changes in the HA and NA of influenza viruses caused by antigenic drift result in the creation of novel strains that the immune system of humans might not be able to identify.[22] These emerging strains are the influenza virus's evolutionary responses to a potent immunological response across the population. The main cause of influenza recurrence is antigenic drift, which makes it essential to reevaluate and update the influenza vaccine's ingredient list every year.[22] Annual influenza outbreaks are caused by antigenic drift and declining immunity, when the residual defenses from prior exposures to related viruses are incomplete. Antigenic drift occurs in Influenza A, B, and C.[22]
Hemagglutination-inhibition experiments using ferret serum after infection allowed the identification of two very different antigenic influenza type B variants in the years 1988 – 1989. These viruses shared antigens with either B/Yamagata/16/88, a variation that was discovered in Japan in May 1988, or B/Victoria/2/87, the most recent reference strain.[32] The B/Victoria/2/87 virus shared antigens with all influenza B viruses discovered in the United States during an outbreak in the winter of 1988 – 1989.[32]
In Japan, influenza B virus reinfection was investigated virologically in 1985 – 1991 and epidemiologically in 1979 – 1991 in children.[33] Four influenza B virus outbreaks that each included antigenic drift occurred during the course of this study. Between the epidemics in 1987 – 1988 and 1989 – 1990, there was a significant genetic and antigenic change in the viruses.[33] Depending on the influenza seasons, the minimum rate of reinfection with influenza B virus for the entire period was between 2 and 25%.[33] Haemagglutination inhibition assays were used to examine the antigens of the influenza B virus primary and reinfection strains that were isolated from 18 children between the years of 1985 and 1990, which encompassed three epidemic periods. The findings revealed that reinfection occurred with the viruses recovered during the 1984 – 1985 and 1987 – 1988 influenza seasons, which belonged to the same lineage and were antigenically close.[33]
Today, reinfection continues as increased influenza activity was reported in the United States in November 2022 due to the winter weather. [34]
References
- Fenner F (1976). "Classification and nomenclature of viruses. Second report of the International Committee on Taxonomy of Viruses" (PDF). Intervirology. 7 (1–2): 1–115. doi:10.1159/000149938. PMID 826499.
- Murphy FA, Fauquet CM, Bishop DH, Ghabrial SA, Jarvis AW, Martelli GP, et al., eds. (1995). "Virus taxonomy: Sixth report of the International Committee on Taxonomy of Viruses" (PDF). Archives of Virology. 10: 350–354.
- Smith GJ, Bahl J, Donis R, Hongo S, Kochs G, Lamb B, et al. (8 June 2017). "Changing individual genus and species names in the family Orthomyxoviridae". International Committee on Taxonomy of Viruses (ICTV). Retrieved 22 March 2019.
- Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y (March 2018). "Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes". Journal of Virology. 92 (6): 561–574. doi:10.1016/B978-0-12-809633-8.21505-7. ISBN 9780128145166. PMC 7268205. PMID 29321324.
- Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA (May 2000). "Influenza B virus in seals". Science. 288 (5468): 1051–1053. Bibcode:2000Sci...288.1051O. doi:10.1126/science.288.5468.1051. PMID 10807575.
- Hay AJ, Gregory V, Douglas AR, Lin YP (December 2001). "The evolution of human influenza viruses". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 356 (1416): 1861–1870. doi:10.1098/rstb.2001.0999. PMC 1088562. PMID 11779385.
- Matsuzaki Y, Sugawara K, Takashita E, Muraki Y, Hongo S, Katsushima N, et al. (September 2004). "Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community". Journal of Medical Virology. 74 (1): 132–140. doi:10.1002/jmv.20156. PMID 15258979. S2CID 31146117.
- Lindstrom SE, Hiromoto Y, Nishimura H, Saito T, Nerome R, Nerome K (May 1999). "Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes". Journal of Virology. 73 (5): 4413–4426. doi:10.1128/JVI.73.5.4413-4426.1999. PMC 104222. PMID 10196339.
- Klimov AI, Garten R, Russell C, Barr IG, Besselaar TG, Daniels R, et al. (October 2012). "WHO recommendations for the viruses to be used in the 2012 Southern Hemisphere Influenza Vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from February to September 2011". Vaccine. 30 (45): 6461–6471. doi:10.1016/j.vaccine.2012.07.089. PMC 6061925. PMID 22917957.
- Moa AM, Chughtai AA, Muscatello DJ, Turner RM, MacIntyre CR (July 2016). "Immunogenicity and safety of inactivated quadrivalent influenza vaccine in adults: A systematic review and meta-analysis of randomised controlled trials". Vaccine. 34 (35): 4092–4102. doi:10.1016/j.vaccine.2016.06.064. PMID 27381642.
- Yamashita M, Krystal M, Fitch WM, Palese P (March 1988). "Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses". Virology. 163 (1): 112–122. doi:10.1016/0042-6822(88)90238-3. PMID 3267218.
- Nobusawa E, Sato K (April 2006). "Comparison of the mutation rates of human influenza A and B viruses". Journal of Virology. 80 (7): 3675–3678. doi:10.1128/JVI.80.7.3675-3678.2006. PMC 1440390. PMID 16537638.
- van de Sandt CE, Bodewes R, Rimmelzwaan GF, de Vries RD (September 2015). "Influenza B viruses: not to be discounted". Future Microbiology. 10 (9): 1447–1465. doi:10.2217/fmb.15.65. PMID 26357957.
- Alhoufie ST, Alsharif NH, Alfarouk KO, Ibrahim NA, Kheyami AM, Aljifri AA (November 2021). "COVID-19 with underdiagnosed influenza B and parainfluenza-2 co-infections in Saudi Arabia: Two case reports". Journal of Infection and Public Health. 14 (11): 1567–1570. doi:10.1016/j.jiph.2021.09.005. PMC 8442300. PMID 34627054.
- Koutsakos M, Wheatley AK, Laurie K, Kent SJ, Rockman S (December 2021). "Influenza lineage extinction during the COVID-19 pandemic?". Nature Reviews. Microbiology. 19 (12): 741–742. doi:10.1038/s41579-021-00642-4. PMC 8477979. PMID 34584246.
- World Health Organization (29 September 2023). "Questions and Answers: Recommended composition of influenza virus vaccines for use in the southern hemisphere 2024 influenza season and development of candidate vaccine viruses for pandemic preparedness" (PDF). Retrieved 26 October 2023.
- Schnirring L (29 September 2023). "WHO advisers recommend switch back to trivalent flu vaccines". CIDRAP. Retrieved 26 October 2023.
- Büchen-Osmond, C. (Ed) (2006). "ICTVdB Virus Description—00.046.0.04. Influenzavirus B". ICTVdB—The Universal Virus Database, version 4. New York: Columbia University. Retrieved 2007-09-15.
- Suzuki Y, Nei M (April 2002). "Origin and evolution of influenza virus hemagglutinin genes". Molecular Biology and Evolution. 19 (4): 501–509. doi:10.1093/oxfordjournals.molbev.a004105. PMID 11919291.
- Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, et al. (April 2018). "The evolutionary history of vertebrate RNA viruses". Nature. 556 (7700): 197–202. Bibcode:2018Natur.556..197S. doi:10.1038/s41586-018-0012-7. PMID 29618816. S2CID 4608233.
- Parry R, Wille M, Turnbull OM, Geoghegan JL, Holmes EC (September 2020). "Divergent Influenza-Like Viruses of Amphibians and Fish Support an Ancient Evolutionary Association". Viruses. 12 (9): 1042. doi:10.3390/v12091042. PMC 7551885. PMID 32962015.
- "Pinkbook: Influenza". U.S. Centers for Disease Control and Prevention (CDC). 2022-09-22. Retrieved 2022-11-23.
- "Influenza Historic Timeline | Pandemic Influenza (Flu)". U.S. Centers for Disease Control and Prevention (CDC). 2022-07-08. Retrieved 2022-11-23.
- "History of influenza vaccination". www.who.int. Retrieved 2022-11-23.
- "When was the Flu Vaccine Invented?". Families Fighting Flu. 2022-02-17. Retrieved 2022-11-23.
- "Quadrivalent Influenza Vaccine". U.S. Centers for Disease Control and Prevention (CDC). 2022-08-25. Retrieved 2022-11-23.
- CDC (2022-11-03). "Selecting Viruses for the Seasonal Flu Vaccine". Centers for Disease Control and Prevention. Retrieved 2022-11-23.
- Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y (March 2018). "Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes". Journal of Virology. 92 (6): 561–574. doi:10.1016/B978-0-12-809633-8.21505-7. ISBN 9780128145166. PMC 7268205. PMID 29321324.
- Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y (March 2018). "Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes". Journal of Virology. 92 (6): 561–574. doi:10.1016/B978-0-12-809633-8.21505-7. ISBN 9780128145166. PMC 7268205. PMID 29321324.
- "Influenza Historic Timeline | Pandemic Influenza (Flu) | CDC". www.cdc.gov. 2022-07-08. Retrieved 2022-11-23.
- Khanmohammadi S, Rezaei N (2022). "Influenza Viruses". Encyclopedia of Infection and Immunity: 67–78. doi:10.1016/B978-0-12-818731-9.00176-2. ISBN 9780323903035. S2CID 239753559.
- Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K (March 1990). "Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983". Virology. 175 (1): 59–68. doi:10.1016/0042-6822(90)90186-u. PMID 2309452.
- Nakajima S, Nishikawa F, Nakamura K, Nakao H, Nakajima K (August 1994). "Reinfection with influenza B virus in children: analysis of the reinfection influenza B viruses". Epidemiology and Infection. 113 (1): 103–112. doi:10.1017/s0950268800051517. PMC 2271217. PMID 8062866.
- "Influenza Situation Report - PAHO/WHO | Pan American Health Organization". www.paho.org. Retrieved 2022-11-23.
External links
- Influenza Research Database Database of influenza genomic sequences and related information.
- Viralzone: Influenzavirus B
- Biere B, Bauer B, Schweiger B (April 2010). "Differentiation of influenza B virus lineages Yamagata and Victoria by real-time PCR". Journal of Clinical Microbiology. 48 (4): 1425–1427. doi:10.1128/JCM.02116-09. PMC 2849545. PMID 20107085.