Tupanvirus
Tupanvirus is a genus of viruses first described in 2018.[1] The genus is composed of two species of virus that are in the giant virus group.[2][3] Researchers discovered the first isolate in 2012 from deep water sediment samples taken at 3000m depth off the coast of Brazil.[1] The second isolate was collected from a soda lake in Southern Nhecolândia, Brazil in 2014.[1] They are named after Tupã (Tupan), a Guaraní thunder god, and the places they were found. These are the first viruses reported to possess genes for amino-acyl tRNA synthetases for all 20 standard amino acids.[1][4][5]
| Tupanvirus | |
|---|---|
 | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Varidnaviria |
| Kingdom: | Bamfordvirae |
| Phylum: | Nucleocytoviricota |
| Class: | Megaviricetes |
| Order: | Imitervirales |
| Family: | Mimiviridae (?) |
| Genus: | Tupanvirus |
| Species | |
| |
Classification
The genus Tupanvirus was first described in 2018 with the discovery of the two isolates of tupanviruses found in soda lake and deep oceanic sediments samples collected in Brazil.[1] The genus is currently unassigned but hypothesized to be a member of the family Mimiviridae, along with the other amoeba-infecting viruses.[6] Members of the family Mimiviridae includes Acanthamoeba polyphaga mimivirus (APMV) that is known for its large size of ~500 nm in diameter.[6] In contrast, the typical virus size range is approximately 20-200 nm.[7] Mimiviruses also possess fibril structures on the capsid as well as genes encoding proteins for nucleotide synthesis and various metabolisms that are not found in other viruses.[8]
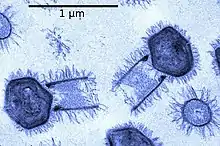
Unlike the other mimiviruses in the Mimiviridae, tupanvirus has a ~550 nm long cylindrical tail covered with fibrils that is attached to the base of the capsid.[6] This morphological feature makes tupanvirus the largest described virus (approximately ~ 1.2 μm in length with the tail) with the longest tail ever observed a in virus.[1][6] Tupanviruses are also capable of infecting a wider range of amoebae than other mimiviruses, and produce a cytotoxic effect in host and non-host organisms that is not observed in APMV.[6] In addition, phylogenomic results revealed that the genus Tupanvirus is separate from the other viruses in the family Mimiviridae.[6]
Morphology
The morphology of Tupanvirus can be similarly compared to that of other ‘amoeba-infecting mimiviruses’.[6] This is because of similarities between the capsid of Tupanvirus and that of other amoebae-infecting mimiviruses.[1] The capsid of Tupanvirus measures approximately 450 nm.[6] One major difference is that the Tupanvirus virion presents a large cylindrical tail (~550 nm × 450 nm diameter) attached to the base of the capsid.[1] Both the capsid and cylindrical tail are covered in fibrils.[6] Tupanvirus can measure up to 1.2 μm in length, and some particles can reach up to 2.3 μm because of the “high degree of plasticity in the size of the tail”.[1][6] Also to note is a lipid membrane is seen within the capsid. Additionally, their capsid is described as having a ‘stargate structure’.[6] This star-shaped vertex is present in other mimivurises, and acts as a seal for the apex of the capsid.[9] It has also been stated that the tail of Tupanvirus is less electron dense than the capsid.[6]
Genome
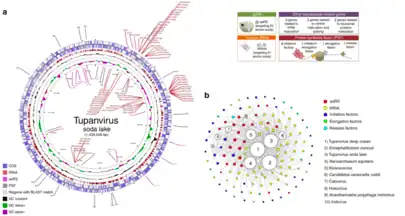
The genome contains roughly 1.5 million base pairs of double-stranded DNA, coding for 1276–1425 predicted proteins, making it the fourth largest among viral genomes.[4] While 30% of the genes are new and not found in other viruses, genomic analysis shows most of the known genes can be related to amoeba-infecting mimiviruses, with the rest corresponding to eukaryotes and bacteria. The shared genes of tupanvirus with different lineages of amoeba-infecting mimiviruses indicate it as a separate genus within family Mimiviridae.[1] For example, its A/T rich genome resembles that of other amoebal mimiviruses and suggests a preference for codons formed by A/T-rich sequences. As well, the frequent occurrence of the “AAAATTGA” promoter motif is also similar to that of other mimiviruses.[1][10]
As a giant virus, tupanvirus presents the largest translational apparatus within the known virosphere, carrying 20 aminoacyl tRNA synthetase (aaRS) and 70 transfer RNAs (tRNA), while the rest are involved in RNA maturation and splicing, as well as ribosomal protein modification.[1] Moreover, tupanviruses contain a number of DNA-independent RNA synthesizing polymerases and enzymes as well as transcription factors that are involved in viral transcription.[10] Furthermore, many genes that encode for processes found in cellular organisms are also found in the Tupanvirus genome, which contains a richer gene set than some bacteria and archaea, and even some eukaryotes.[1] As a result, the analysis of tupanviruses constitutes a new step towards understanding the evolution of giant viruses, because such diverse and complete gene sets not only surpass that of other viral genomes, but also rival that of bacteria and even eukaryotes. In fact, tupanvirus is the only known virus to host all 20 aaRS, exceeding other giant viruses like Klosneuvirus; yet, there is no agreement on whether these genes are host-derived or passed down from an ancestral mimivirus.[6] One such extraordinary example are two copies of an 18 S rRNA intronic region that are highly expressed during viral replication. Although these intronic regions also exist in other mimiviruses, the tupanvirus 18S rRNA is phylogenetically different with their function still unknown.[1]
Host
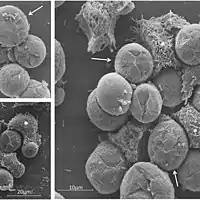
Tupanviruses have the ability to infect a wider range of hosts than other giant viruses, including many amoebae of the genus Acanthameoba (as well as members of the species Vermamoeba vermiformis, Dictyostelium discoideum and Willartia magna),[6] and may also infect other protists. There are no documented threats to humans.[4] The tupanvirus’ generalist approach may be attributed to the low species richness and abundance of the virus’ habitat.[12]
Tupanvirus-infected amoebas also present a new virus-host interaction not previously observed among other amoebae. Infected cells were found to aggregate with non-infected cells, a mechanism suggested to be mediated by mannose binding protein gene expression (MBP).[11] The clusters of affected cells were shown to increase viral dissemination within the host, thus increasing virulence of the virus.[11] The generalist approach, in conjunction with aggregation behaviour, decreases the dilution effect and increases host-encounter rates.
Life cycle and host interaction
Attachment
Viral particles attach directly to the host cell surface.[11] The attachment process occurs very rapidly. Virions can be visibly attached at 0 hours past infection. The specific mechanism is still unknown.[11]
Entry
Virions enter via phagocytosis.[13] Generally only one particle will be present in each phagosome, although several particles may enter the intracellular matrix in different phagosomes at the same time.[13] This Tupanvirus uses membrane fusion in order to release its genome. The viral capsid contains a lipid membrane that facilitates fusion with the phagosome membrane to release the viral genome. The virus also releases the contents kept in the tail after an invagination of the phagosome between the two tail components results in fusion.[1]
Replication
The viral genome is released from the phagosome into the amoeba’s cytoplasm.[1] This virus facilitates aggregation of amoeba hosts in order to replicate rapidly and have a supply of hosts for progeny particles.[11] Bunch formation happens rapidly after infection and will continue as long as host cells are living. Bunches can re-form after mechanical separation as long as host cells are living.[11] Replication factories are formed as early as 8 h.p.i. up to 12 h.p.i.[11][13]
Assembly
Initially, when replication factories are still immature, new virions are assembled as soon as possible.[13] Therefore, many particles at various stages of assembly may be present inside a single factory. Once the factory has matured capsid assembly is finished and the genome is incorporated. The tails are attached to the capsid following genome incorporation.[13] Assembly of particles happens on a loose timeline, resulting in particles at varying stages of assembly at the time of release.
Release
Viral particles are released, facilitated by cell lysis.[13] When infecting the amoeba in the species Vermamoeba vermiformis many assembled particles are not functional at the time of release. As many as half of released particles are not infectious; this phenomenon is also present when infecting host amoeba Acanthamoeba castellanii.[13] Although more research is required it is hypothesized that the production of non-infectious particles may be a normal part of the replication cycle.[13]
References
- Abrahão J, Silva L, Silva LS, Khalil JY, Rodrigues R, Arantes T, et al. (February 2018). "Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere". Nature Communications. 9 (1): 749. Bibcode:2018NatCo...9..749A. doi:10.1038/s41467-018-03168-1. PMC 5829246. PMID 29487281.
- "Taxon Details | ICTV". ictv.global. Retrieved 2023-09-19.
- "Taxon Details | ICTV". ictv.global. Retrieved 2023-09-19.
- Garisto D (27 February 2018). "These giant viruses have more protein-making gear than any known virus". Science News. Retrieved 2021-04-07.
- Schulz F (2017-04-07). "Giant viruses with an expanded complement of translation system components". Science. eScholarship, University of California. 356 (6333): 82–85. Bibcode:2017Sci...356...82S. doi:10.1126/science.aal4657. OCLC 1114659455. PMID 28386012.
- Rodrigues RA, Mougari S, Colson P, La Scola B, Abrahão JS (January 2019). ""Tupanvirus", a new genus in the family Mimiviridae". Archives of Virology. 164 (1): 325–331. doi:10.1007/s00705-018-4067-4. PMID 30291500.
- Fuhrman JA (June 1999). "Marine viruses and their biogeochemical and ecological effects". Nature. 399 (6736): 541–8. Bibcode:1999Natur.399..541F. doi:10.1038/21119. PMID 10376593.
- Colson P, La Scola B, Levasseur A, Caetano-Anollés G, Raoult D (April 2017). "Mimivirus: leading the way in the discovery of giant viruses of amoebae". Nature Reviews. Microbiology. 15 (4): 243–254. doi:10.1038/nrmicro.2016.197. PMC 7096837. PMID 28239153.
- Pires de Souza GA, Rolland C, Nafeh B, La Scola B, Colson P (April 2021). "Giant virus-related sequences in the 5300-year-old Ötzi mummy metagenome". Virus Genes. 57 (2): 222–227. doi:10.1007/s11262-021-01823-2. ISSN 0920-8569. PMID 33566217.
- Rodrigues RA, Arantes TS, Oliveira GP, Dos Santos Silva LK, Abrahão JS (January 2019). Kielian M, Mettenleiter TC, Roossinck MJ (eds.). "The Complex Nature of Tupanviruses". Advances in Virus Research. Academic Press. 103: 135–166. doi:10.1016/bs.aivir.2018.09.001. ISBN 9780128177228. PMID 30635075.
- Oliveira G, Silva L, Leão T, Mougari S, da Fonseca FG, Kroon EG, et al. (January 2019). "Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination". Scientific Reports. 9 (1): 183. Bibcode:2019NatSR...9..183O. doi:10.1038/s41598-018-36552-4. PMC 6336878. PMID 30655573.
- Sergeev VN, Gerasimenko LM, Zavarzin GA (2002-11-01). "[Proterozoic history and present state of cyanobacteria]". Mikrobiologiia. 71 (6): 725–40. doi:10.1023/A:1021415503436. PMID 12526193.
- Silva LC, Rodrigues RA, Oliveira GP, Dornas FP, La Scola B, Kroon EG, Abrahão JS (2019). "Microscopic Analysis of the Tupanvirus Cycle in Vermamoeba vermiformis". Frontiers in Microbiology. 10: 671. doi:10.3389/fmicb.2019.00671. PMC 6456662. PMID 31001237.