Striosome
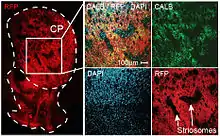
The striosomes (also referred to as striatal patches) are one of two complementary chemical compartments within the striatum (the other compartment is known as the matrix) that can be visualized by staining for immunocytochemical markers such as mu opioid receptors,[1] acetylcholinesterase,[2] enkephalin, substance P, limbic system-associated membrane protein (LAMP),[3] AMPA receptor subunit 1 (GluR1),[4] dopamine receptor subunits, and calcium binding proteins.[5] Striosomal abnormalities have been associated with neurological disorders, such as mood dysfunction in Huntington's disease,[6] though their precise function remains unknown. Recently studies have identified the presence of "exo-patch" neurons that are biochemically and genetically the same as striosomal neurons, but reside in the matrix compartment. [7] This study also characterized the different input and output connections of the striosome and matrix compartments, revealing that both regions have direct inputs to dopamine neurons (though the striosome inputs are somatic whereas the matrix targets distal dendrites). The authors also revealed unique inputs to the striosome from subcortical limbic structures like the amygdala and bed nucleus of the stria terminalis.
| Striosome | |
|---|---|
| Identifiers | |
| NeuroLex ID | nlx_anat_20090506 |
| Anatomical terms of neuroanatomy | |
Striosomes (a.k.a striatal "patches") were discovered by Candace Pert in 1976 based on mu opioid receptor autoradiography and Ann Graybiel in 1978 using acetylcholinesterase histochemistry.
References
- Pert CB, Kuhar MJ, Snyder SH (Oct 1976). "Opiate receptor: autoradiographic localization in rat brain". Proc. Natl. Acad. Sci. 73 (10): 3792–33. doi:10.1073/pnas.73.10.3729. PMC 431193. PMID 185626.
- Graybiel AM, Ragsdale CW Jr (Nov 1978). "Histochemically distinct compartments in the striatum of human, monkeys, and cat demonstrated by acetylthiocholinesterase staining". Proc Natl Acad Sci U S A. 75 (11): 5723–6. doi:10.1073/pnas.75.11.5723. PMC 393041. PMID 103101.
- Prensa L, Giménez-Amaya JM, Parent A (Nov 1999). "Chemical heterogeneity of the striosomal compartment in the human striatum". J Comp Neurol. 413 (4): 603–18. doi:10.1002/(SICI)1096-9861(19991101)413:4<603::AID-CNE9>3.0.CO;2-K. PMID 10495446.
- Martin LJ, Blackstone CD, Huganir RL, Price DL (Feb 1993). "The striatal mosaic in primates: striosomes and matrix are differentially enriched in ionotropic glutamate receptor subunits". J. Neurosci. 13 (2): 782–92. doi:10.1523/JNEUROSCI.13-02-00782.1993. PMC 6576641. PMID 7678861.
- O'Kusky JR, Nasir J, Cicchetti F, Parent A, Hayden MR (Feb 1999). "Neuronal degeneration in the basal ganglia and loss of pallido-subthalamic synapses in mice with targeted disruption of the Huntington's disease gene". Brain Res. 818 (2): 468–79. doi:10.1016/S0006-8993(98)01312-2. PMID 10082833. S2CID 45823601.
- Tippett LJ, Waldvogel HJ, Thomas SJ, Hogg VM, van Roon-Mom W, Synek BJ, Graybiel AM, Faull RL (Jan 2007). "Striosomes and mood dysfunction in Huntington's disease". Brain. 130 (1): 206–21. doi:10.1093/brain/awl243. PMID 17040921.
- Smith JB, Klug JR, Ross DL, Howard CD, Hollon NG, Ko VI, Hoffman H, Callaway EM, Gerfen CR, Jin X (Sep 2016). "Genetic-Based Dissection Unveils the Inputs and Outputs of Striatal Patch and Matrix Compartments". Neuron. 91 (5): 1069–84. doi:10.1016/j.neuron.2016.07.046. PMID 27568516.
- Reinius B; et al. (March 27, 2015). "Conditional targeting of medium spiny neurons in the striatal matrix". Front. Behav. Neurosci. 9: 71. doi:10.3389/fnbeh.2015.00071. PMC 4375991. PMID 25870547.
External links
- Trafton, Anne (2020-10-27). "Study helps explain why motivation to learn declines with age". MIT News | Massachusetts Institute of Technology. Retrieved 2020-10-29. Article on striosomes' role in age-related decline in learning.