Short interspersed nuclear element
Short interspersed nuclear elements (SINEs) are non-autonomous, non-coding transposable elements (TEs) that are about 100 to 700 base pairs in length.[1] They are a class of retrotransposons, DNA elements that amplify themselves throughout eukaryotic genomes, often through RNA intermediates. SINEs compose about 13% of the mammalian genome.[2]
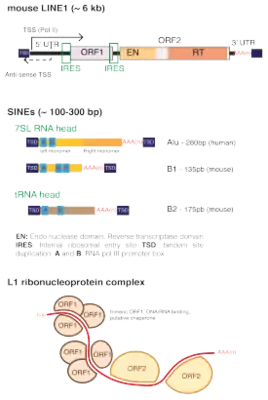
The internal regions of SINEs originate from tRNA and remain highly conserved, suggesting positive pressure to preserve structure and function of SINEs.[3] While SINEs are present in many species of vertebrates and invertebrates, SINEs are often lineage specific, making them useful markers of divergent evolution between species. Copy number variation and mutations in the SINE sequence make it possible to construct phylogenies based on differences in SINEs between species. SINEs are also implicated in certain types of genetic disease in humans and other eukaryotes.
In essence, short interspersed nuclear elements are genetic parasites which have evolved very early in the history of eukaryotes to utilize protein machinery within the organism as well as to co-opt the machinery from similarly parasitic genomic elements. The simplicity of these elements make them remarkably successful at persisting and amplifying (through retrotransposition) within the genomes of eukaryotes. These "parasites" which have become ubiquitous in genomes can be very deleterious to organisms as discussed below. However, eukaryotes have been able to integrate short-interspersed nuclear elements into different signaling, metabolic and regulatory pathways and SINEs have become a great source of genetic variability. They seem to play a particularly important role in the regulation of gene expression and the creation of RNA genes. This regulation extends to chromatin re-organization and the regulation of genomic architecture. The different lineages, mutations, and activities among eukaryotes make short-interspersed nuclear elements a useful tool in phylogenetic analysis.
Classification and structure
SINEs are classified as non-LTR retrotransposons because they do not contain long terminal repeats (LTRs).[4] There are three types of SINEs common to vertebrates and invertebrates: CORE-SINEs, V-SINEs, and AmnSINEs.[3] SINEs have 50-500 base pair internal regions which contain a tRNA-derived segment with A and B boxes that serve as an internal promoter for RNA polymerase III.[5][3]
Internal structure
SINEs are characterized by their different modules, which are essentially a sectioning of their sequence. SINEs can, but do not necessarily have to possess a head, a body, and a tail. The head, is at the 5' end of short-interspersed nuclear elements and is an evolutionarily derived from an RNA synthesized by RNA Polymerase III such as ribosomal RNAs and tRNAs; the 5' head is indicative of which endogenous element that SINE was derived from and was able to parasitically utilize its transcriptional machinery.[1] For example, the 5' of the Alu sine is derived from 7SL RNA, a sequence transcribed by RNA Polymerase III which codes for the RNA element of SRP, an abundant ribonucleoprotein.[6] The body of SINEs possess an unknown origin but often share much homology with a corresponding LINE which thus allows SINEs to parasitically co-opt endonucleases coded by LINEs (which recognize certain sequence motifs). Lastly, the 3′ tail of SINEs is composed of short simple repeats of varying lengths; these simple repeats are sites where two (or more) short-interspersed nuclear elements can combine to form a dimeric SINE.[7] Short-interspersed nuclear elements which only possess a head and tail are called simple SINEs whereas short-interspersed nuclear elements which also possess a body or are a combination of two or more SINEs are complex SINEs.[1]
Transcription
Short-interspersed nuclear elements are transcribed by RNA polymerase III which is known to transcribe ribosomal RNA and tRNA, two types of RNA vital to ribosomal assembly and mRNA translation.[8] SINEs, like tRNAs and many small-nuclear RNAs possess an internal promoter and thus are transcribed differently than most protein-coding genes.[1] In other words, short-interspersed nuclear elements have their key promoter elements within the transcribed region itself. Though transcribed by RNA polymerase III, SINEs and other genes possessing internal promoters, recruit different transcriptional machinery and factors than genes possessing upstream promoters.[9]
Effects on gene expression
Changes in chromosome structure influence gene expression primarily by affecting the accessibility of genes to transcriptional machinery. The chromosome has a very complex and hierarchical system of organizing the genome. This system of organization, which includes histones, methyl groups, acetyl groups, and a variety of proteins and RNAs allows different domains within a chromosome to be accessible to polymerases, transcription factors, and other associated proteins to different degrees.[10] Furthermore, the shape and density of certain areas of a chromosome can affect the shape and density of neighboring (or even distant regions) on the chromosome through interaction facilitated by different proteins and elements. Non-coding RNAs such as short-interspersed nuclear elements, which have been known to associate with and contribute to chromatin structure, can thus play huge role in regulating gene expression.[11] Short-interspersed-nuclear-elements similarly can be involved in gene regulation by modifying genomic architecture.
In fact Usmanova et al. 2008 suggested that short-interspersed nuclear elements can serve as direct signals in chromatin rearrangement and structure. The paper examined the global distribution of SINEs in mouse and human chromosomes and determined that this distribution was very similar to genomic distributions of genes and CpG motifs.[12] The distribution of SINEs to genes was significantly more similar than that of other non-coding genetic elements and even differed significantly from the distribution of long-interspersed nuclear elements.[12] This suggested that the SINE distribution was not a mere accident caused by LINE-mediated retrotransposition but rather that SINEs possessed a role in gene-regulation. Furthermore, SINEs frequently contain motifs for YY1 polycomb proteins.[12] YY1 is a zinc-finger protein that acts as a transcriptional repressor for a wide-variety of genes essential for development and signaling.[13] Polycomb protein YY1 is believed to mediate the activity of histone deacetylases and histone acetyltransferases to facilitate chromatin re-organization; this is often to facilitate the formation of heterochromatin (gene-silencing state).[14] Thus, the analysis suggests that short-interspersed nuclear elements can function as a ‘signal-booster' in the polycomb-dependent silencing of gene-sets through chromatin re-organization.[12] In essence, it is the cumulative effect of many types of interactions that leads to the difference between euchromatin, which is not tightly packed and generally more accessible to transcriptional machinery, and heterochromatin, which is tightly packed and generally not accessible to transcriptional machinery; SINEs seem to play an evolutionary role in this process.
In addition to directly affecting chromatin structure, there are a number of ways in which SINEs can potentially regulate gene expression. For example, long non-coding RNA can directly interact with transcriptional repressors and activators, attenuating or modifying their function.[15] This type of regulation can occur in different ways: the RNA transcript can directly bind to the transcription factor as a co-regulator; also, the RNA can regulate and modify the ability of co-regulators to associate with the transcription factor.[15] For example, Evf-2, a certain long non-coding RNA, has been known to function as a co-activator for certain homeobox transcription factors which are critical to nervous system development and organization.[16] Furthermore, RNA transcripts can interfere with the functionality of the transcriptional complex by interacting or associating with RNA polymerases during the transcription or loading processes.[15] Moreover, non-coding RNAs like SINEs can bind or interact directly with the DNA duplex coding the gene and thus prevent its transcription.[15]
Also, many non-coding RNAs are distributed near protein-coding genes, often in the reverse direction. This is especially true for short-interspersed nuclear elements as seen in Usmanova et al. These non-coding RNAs, which lie adjacent to or overlap gene-sets provide a mechanism by which transcription factors and machinery can be recruited to increase or repress the transcription of local genes. The particular example of SINEs potentially recruiting the YY1 polycomb transcriptional repressor is discussed above.[12] Alternatively, it also provides a mechanism by which local gene expression can be curtailed and regulated because the transcriptional complexes can hinder or prevent nearby genes from being transcribed. There is research to suggest that this phenomenon is particularly seen in the gene-regulation of pluripotent cells.[17]
In conclusion, non-coding RNAs such as SINEs are capable of affecting gene expression on a multitude of different levels and in different ways. Short-interspersed nuclear elements are believed to be deeply integrated into a complex regulatory network capable of fine-tuning gene expression across the eukaryotic genome.
Propagation and regulation
The RNA coded by the short-interspersed nuclear element does not code for any protein product but is nonetheless reverse-transcribed and inserted back into an alternate region in the genome. For this reason, short interspersed nuclear elements are believed to have co-evolved with long interspersed nuclear element (LINEs), as LINEs do in fact encode protein products which enable them to be reverse- transcribed and integrated back into the genome.[4] SINEs are believed to have co-opted the proteins coded by LINEs which are contained in 2 reading frames. Open reading frame 1 (ORF 1) encodes a protein which binds to RNA and acts as a chaperone to facilitate and maintain the LINE protein-RNA complex structure.[18] Open reading frame 2 (ORF 2) codes a protein which possesses both endonuclease and reverse transcriptase activities.[19] This enables the LINE mRNA to be reverse-transcribed into DNA and integrated into the genome based on the sequence-motifs recognized by the protein's endonuclease domain.
LINE-1 (L1) is transcribed and retrotransposed most frequently in the germ-line and during early development; as a result SINEs move around the genome most during these periods. SINE transcription is down-regulated by transcription factors in somatic cells after early development, though stress can cause up-regulation of normally silent SINEs.[20] SINEs can be transferred between individuals or species via horizontal transfer through a viral vector.[21]
SINEs are known to share sequence homology with LINES which gives a basis by which the LINE machinery can reverse transcribe and integrate SINE transcripts.[22] Alternately, some SINEs are believed to use a much more complex system of integrating back into the genome; this system involves the use random double-stranded DNA breaks (rather than the endonuclease coded by related long-interspersed nuclear elements creating an insertion-site).[22] These DNA breaks are utilized to prime reverse transcriptase, ultimately integrating the SINE transcript back into the genome.[22] SINEs nonetheless depend on enzymes coded by other DNA elements and are thus known as non-autonomous retrotransposons as they depend on the machinery of LINEs, which are known as autonomous retrotransposons.<[23]
The theory that short-interspersed nuclear elements have evolved to utilize the retrotransposon machinery of long-interspersed nuclear elements is supported by studies which examine the presence and distribution of LINEs and SINEs in taxa of different species.[24] For example, LINEs and SINEs in rodents and primates show very strong homology at the insertion-site motif.[24] Such evidence is a basis for the proposed mechanism in which integration of the SINE transcript can be co-opted with LINE-coded protein products. This is specifically demonstrated by a detailed analysis of over 20 rodent species profiled LINEs and SINEs, mainly L1s and B1s respectively; these are families of LINEs and SINEs found at high frequencies in rodents along with other mammals.[24] The study sought to provide phylogenetic clarity within the context of LINE and SINE activity.
The study arrived at a candidate taxa believed to be the first instance of L1 LINE extinction; it expectedly discovered that there was no evidence to suggest that B1 SINE activity occurred in species which did not have L1 LINE activity.[24] Also, the study suggested that B1 short-interspersed nuclear element silencing in fact occurred before L1 long-interspersed nuclear element extinction; this is due to the fact that B1 SINEs are silenced in the genus most-closely related to the genus which does not contain active L1 LINEs (though the genus with B1 SINE silencing still contains active L1 LINEs).[24] Another genus was also found which similarly contained active L1 long-interspersed nuclear elements but did not contain B1 short-interspersed nuclear elements; the opposite scenario, in which active B1 SINEs were present in a genus which did not possess active L1 LINEs was not found.[24] This result was expected and strongly supports the theory that SINEs have evolved to co-opt the RNA-binding proteins, endonucleases, and reverse-transcriptases coded by LINEs. In taxa which do not actively transcribe and translate long-interspersed nuclear elements protein-products, SINEs do not have the theoretical foundation by which to retrotranspose within the genome. The results obtained in Rinehart et al. are thus very supportive of the current model of SINE retrotransposition.
Effects of SINE transposition
Insertion of a SINE upstream of a coding region may result in exon shuffling or changes to the regulatory region of the gene. Insertion of a SINE into the coding sequence of a gene can have deleterious effects and unregulated transposition can cause genetic disease. The transposition and recombination of SINEs and other active nuclear elements is thought to be one of the major contributions of genetic diversity between lineages during speciation.[21]
Common SINEs
Short-interspersed nuclear elements are believed to have parasitic origins in eukaryotic genomes. These SINEs have mutated and replicated themselves a large number of times on an evolutionary time-scale and thus form many different lineages. Their early evolutionary origin has caused them to be ubiquitous in many eukaryotic lineages.
Alu elements, short-interspersed nuclear element of about 300 nucleotides, are the most common SINE in humans, with >1,000,000 copies throughout the genome, which is over 10 percent of the total genome; this is not uncommon among other species.[25] Alu element copy number differences can be used to distinguish between and construct phylogenies of primate species.[21] Canines differ primarily in their abundance of SINEC_Cf repeats throughout the genome, rather than other gene or allele level mutations. These dog-specific SINEs may code for a splice acceptor site, altering the sequences that appear as exons or introns in each species.[26]
Apart from mammals, SINEs can reach high copy numbers in a range of species, including nonbony vertebrates (elephant shark) and some fish species (coelacanths).[27] In plants, SINEs are often restricted to closely related species and have emerged, decayed, and vanished frequently during evolution.[28] Nevertheless, some SINE families such as the Au-SINEs[29] and the Angio-SINEs[30] are unusually widespread across many often unrelated plant species.
Diseases
There are >50 human diseases associated with SINEs.[20] When inserted near or within the exon, SINEs can cause improper splicing, become coding regions, or change the reading frame, often leading to disease phenotypes in humans and other animals.[26] Insertion of Alu elements in the human genome is associated with breast cancer, colon cancer, leukemia, hemophilia, Dent's disease, cystic fibrosis, neurofibromatosis, and many others.[4]
microRNAs
The role of short-interspersed nuclear elements in gene regulation within cells has been supported by multiple studies. One such study examined the correlation between a certain family of SINEs with microRNAs (in zebrafish).[31] The specific family of SINEs being examined was the Anamnia V-SINEs; this family of short interspersed nuclear elements is often found in the untranslated region of the 3' end of many genes and is present in vertebrate genomes.[31] The study involved a computational analysis in which the genomic distribution and activity of the Anamnia V-SINEs in Danio rerio zebrafish was examined; furthermore, these V-SINEs potential to generate novel microRNA loci was analyzed.[31] It was found that genes which were predicted to possess V-SINEs were targeted by microRNAs with significantly higher hybridization E-values (relative to other areas in the genome).[31] The genes that had high hybridization E-values were genes particularly involved in metabolic and signaling pathways.[31] Almost all miRNAs identified to have a strong ability to hybridize to putative V-SINE sequence motifs in genes have been identified (in mammals) to have regulatory roles.[31] These results which establish a correlation between short-interspersed nuclear elements and different regulatory microRNAs strongly suggest that V-SINEs have a significant role in attenuating responses to different signals and stimuli related to metabolism, proliferation and differentiation. Many other studies must be undertaken to establish the validity and extent of short-interspersed nuclear element retrotransposons' role in regulatory gene-expression networks. In conclusion, though not much is known about the role and mechanism by which SINEs generate miRNA gene loci it is generally understood that SINEs have played a significant evolutionary role in the creation of "RNA-genes", this is also touched upon above in SINEs and pseudogenes.
With such evidence suggesting that short-interspersed nuclear elements have been evolutionary sources for microRNA loci generation it is important to further discuss the potential relationships between the two as well as the mechanism by which the microRNA regulates RNA degradation and more broadly, gene expression. A microRNA is a non-coding RNA generally 22 nucleotides in length.[32] This non-protein coding oligonucleotide is itself coded by longer nuclear DNA sequence usually transcribed by RNA polymerase II which is also responsible for the transcription of most mRNAs and snRNAs in eukaryotes.[33] However, some research suggests that some microRNAs that possess upstream short-interspersed nuclear elements are transcribed by RNA polymerase III which is widely implicated in ribosomal RNA and tRNA, two transcripts vital to mRNA translation.[34] This provides an alternate mechanism by which short-interspersed nuclear elements could be interacting with or mediating gene-regulatory networks involving microRNAs.
The regions coding miRNA can be independent RNA-genes often being anti-sense to neighboring protein-coding genes, or can be found within the introns of protein-coding genes.[35] The co-localization of microRNA and protein-coding genes provides a mechanistic foundation by which microRNA regulates gene-expression. Furthermore, Scarpato et al. reveals (as discussed above) that genes predicted to possess short-interspersed nuclear elements (SINEs) through sequence analysis were targeted and hybridized by microRNAs significantly greater than other genes.[31] This provides an evolutionarily path by which the parasitic SINEs were co-opted and utilized to form RNA-genes (such as microRNAs) which have evolved to play a role in complex gene-regulatory networks.
The microRNAs are transcribed as part of longer RNA strands of generally about 80 nucleotides which through complementary base-pairing are able to form hairpin loop structures[36] These structures are recognized and processed in the nucleus by the nuclear protein DiGeorge Syndrome Critical Region 8 (DGCR8) which recruits and associates with the Drosha protein.[37] This complex is responsible for cleaving some of the hair-pin structures from the pre-microRNA which is transported to the cytoplasm. The pre-miRNA is processed by the protein DICER into a double stranded 22 nucleotide.[38] Thereafter, one of the strands is incorporated into a multi-protein RNA-induced silencing complex (RISC).[39] Among these proteins are proteins from the Argonaute family which are critical to the complex's ability to interact with and repress the translation of the target mRNA.[40]
Understanding the different ways in which microRNA regulates gene-expression, including mRNA-translation and degradation is key to understanding the potential evolutionary role of SINEs in gene-regulation and in the generation of microRNA loci. This, in addition to SINEs' direct role in regulatory networks (as discussed in SINEs as long non-coding RNAs) is crucial to beginning to understand the relationship between SINEs and certain diseases. Multiple studies have suggested that increased SINE activity is correlated with certain gene-expression profiles and post-transcription regulation of certain genes.[41][42][43] In fact, Peterson et al. 2013 demonstrated that high SINE RNA expression correlates with post-transcriptional downregulation of BRCA1, a tumor suppressor implicated in multiple forms of cancer, namely breast cancer.[43] Furthermore, studies have established a strong correlation between transcriptional mobilization of SINEs and certain cancers and conditions such as hypoxia; this can be due to the genomic instability caused by SINE activity as well as more direct-downstream effects.[42] SINEs have also been implicated in countless other diseases. In essence, short-interspersed nuclear elements have become deeply integrated in countless regulatory, metabolic and signaling pathways and thus play an inevitable role in causing disease. Much is still to be known about these genomic parasites but it is clear they play a significant role within eukaryotic organisms.
SINEs and pseudogenes
The activity of SINEs however has genetic vestiges which do not seem to play a significant role, positive or negative, and manifest themselves in the genome as pseudogenes. SINEs however should not be mistaken as RNA pseudogenes.[1] In general, pseudogenes are generated when processed mRNAs of protein-coding genes are reverse-transcribed and incorporated back into the genome (RNA pseudogenes are reverse transcribed RNA genes).[44] Pseudogenes are generally functionless as they descend from processed RNAs independent of their evolutionary-context which includes introns and different regulatory elements which enable transcription and processing. These pseudogenes, though non-functional may in some cases still possess promoters, CpG islands, and other features which enable transcription; they thus can still be transcribed and may possess a role in the regulation of gene expression (like SINEs and other non-coding elements).[44] Pseudogenes thus differ from SINEs in that they are derived from transcribed- functional RNA whereas SINEs are DNA elements which retrotranspose by co-opting RNA genes transcriptional machinery. However, there are studies which suggest that retro-transposable elements such as short-interspersed nuclear elements are not only capable of copying themselves in alternate regions in the genome but are also able to do so for random genes too.[45][46] Thus SINEs can be playing a vital role in the generation of pseudogenes, which themselves are known to be involved in regulatory networks. This is perhaps another means by which SINEs have been able to influence and contribute to gene-regulation.
References
- Vassetzky NS, Kramerov DA (January 2013). "SINEBase: a database and tool for SINE analysis". Nucleic Acids Research. 41 (Database issue): D83-9. doi:10.1093/nar/gks1263. PMC 3531059. PMID 23203982.
- Ishak, Charles A.; De Carvalho, Daniel D. (2020). "Reactivation of Endogenous Retroelements in Cancer Development and Therapy". Annual Review of Cancer Biology. 4: 159–176. doi:10.1146/annurev-cancerbio-030419-033525.
- Sun FJ, Fleurdépine S, Bousquet-Antonelli C, Caetano-Anollés G, Deragon JM (January 2007). "Common evolutionary trends for SINE RNA structures". Trends in Genetics. 23 (1): 26–33. doi:10.1016/j.tig.2006.11.005. PMID 17126948.
- Hancks DC, Kazazian HH (June 2012). "Active human retrotransposons: variation and disease". Current Opinion in Genetics & Development. 22 (3): 191–203. doi:10.1016/j.gde.2012.02.006. PMC 3376660. PMID 22406018.
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. (December 2007). "A unified classification system for eukaryotic transposable elements". Nature Reviews. Genetics. 8 (12): 973–82. doi:10.1038/nrg2165. PMID 17984973. S2CID 32132898.
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J (April 2007). "Evolutionary history of 7SL RNA-derived SINEs in Supraprimates". Trends in Genetics. 23 (4): 158–61. doi:10.1016/j.tig.2007.02.002. PMID 17307271.
- Okada N, Hamada M, Ogiwara I, Ohshima K (December 1997). "SINEs and LINEs share common 3' sequences: a review". Gene. 205 (1–2): 229–43. doi:10.1016/s0378-1119(97)00409-5. PMID 9461397.
- Deininger PL, Batzer MA (October 2002). "Mammalian retroelements". Genome Research. 12 (10): 1455–65. doi:10.1101/gr.282402. PMID 12368238.
- White RJ (May 2011). "Transcription by RNA polymerase III: more complex than we thought". Nature Reviews. Genetics. 12 (7): 459–63. doi:10.1038/nrg3001. PMID 21540878. S2CID 21123216.
- Kiefer JC (April 2007). "Epigenetics in development". Developmental Dynamics. 236 (4): 1144–56. doi:10.1002/dvdy.21094. PMID 17304537.
- Rodríguez-Campos A, Azorín F (November 2007). "RNA is an integral component of chromatin that contributes to its structural organization". PLOS ONE. 2 (11): e1182. Bibcode:2007PLoSO...2.1182R. doi:10.1371/journal.pone.0001182. PMC 2063516. PMID 18000552.
- Usmanova NM, Kazakov VI, Tomilin NV (2008). "[SINEs in mammalian genomes can serve as additional signals in formation of facultative heterochromatin]". Tsitologiia (in Russian). 50 (3): 256–60. PMID 18664128.
- Shi Y, Seto E, Chang LS, Shenk T (October 1991). "Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein". Cell. 67 (2): 377–88. doi:10.1016/0092-8674(91)90189-6. PMID 1655281. S2CID 19399858.
- Yao YL, Yang WM, Seto E (September 2001). "Regulation of transcription factor YY1 by acetylation and deacetylation". Molecular and Cellular Biology. 21 (17): 5979–91. doi:10.1128/mcb.21.17.5979-5991.2001. PMC 87316. PMID 11486036.
- Goodrich JA, Kugel JF (August 2006). "Non-coding-RNA regulators of RNA polymerase II transcription". Nature Reviews. Molecular Cell Biology. 7 (8): 612–6. doi:10.1038/nrm1946. PMID 16723972. S2CID 22274894.
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD (June 2006). "The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator". Genes & Development. 20 (11): 1470–84. doi:10.1101/gad.1416106. PMC 1475760. PMID 16705037.
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, et al. (May 2016). "Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells". Cell Stem Cell. 18 (5): 637–52. doi:10.1016/j.stem.2016.01.024. PMID 26996597.
- Ewing AD, Ballinger TJ, Earl D, Harris CC, Ding L, Wilson RK, Haussler D (March 2013). "Retrotransposition of gene transcripts leads to structural variation in mammalian genomes". Genome Biology. 14 (3): R22. doi:10.1186/gb-2013-14-3-r22. PMC 3663115. PMID 23497673.
- Mätlik K, Redik K, Speek M (2006). "L1 antisense promoter drives tissue-specific transcription of human genes". Journal of Biomedicine & Biotechnology. 2006 (1): 71753. doi:10.1155/JBB/2006/71753. PMC 1559930. PMID 16877819.
- Beauregard A, Curcio MJ, Belfort M (2008). "The take and give between retrotransposable elements and their hosts". Annual Review of Genetics. 42: 587–617. doi:10.1146/annurev.genet.42.110807.091549. PMC 2665727. PMID 18680436.
- Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN (2008). "Transposable elements as drivers of genomic and biological diversity in vertebrates". Chromosome Research. 16 (1): 203–15. doi:10.1007/s10577-007-1202-6. PMID 18293113. S2CID 10510149.
- Singer MF (March 1982). "SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes". Cell. 28 (3): 433–4. doi:10.1016/0092-8674(82)90194-5. PMID 6280868. S2CID 22129236.
- Gogvadze E, Buzdin A (December 2009). "Retroelements and their impact on genome evolution and functioning". Cellular and Molecular Life Sciences. 66 (23): 3727–42. doi:10.1007/s00018-009-0107-2. PMID 19649766. S2CID 23872541.
- Rinehart TA, Grahn RA, Wichman HA (2005). "SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms". Cytogenetic and Genome Research. 110 (1–4): 416–25. doi:10.1159/000084974. PMID 16093694. S2CID 36518754.
- Cordaux R, Batzer MA (October 2009). "The impact of retrotransposons on human genome evolution". Nature Reviews. Genetics. 10 (10): 691–703. doi:10.1038/nrg2640. PMC 2884099. PMID 19763152.
- Wang W, Kirkness EF (December 2005). "Short interspersed elements (SINEs) are a major source of canine genomic diversity". Genome Research. 15 (12): 1798–808. doi:10.1101/gr.3765505. PMC 1356118. PMID 16339378.
- Chalopin D, Naville M, Plard F, Galiana D, Volff JN (January 2015). "Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates". Genome Biology and Evolution. 7 (2): 567–80. doi:10.1093/gbe/evv005. PMC 4350176. PMID 25577199.
- Kramerov DA, Vassetzky NS (December 2011). "Origin and evolution of SINEs in eukaryotic genomes". Heredity. 107 (6): 487–95. doi:10.1038/hdy.2011.43. PMC 3242629. PMID 21673742.
- Fawcett JA, Kawahara T, Watanabe H, Yasui Y (June 2006). "A SINE family widely distributed in the plant kingdom and its evolutionary history". Plant Molecular Biology. 61 (3): 505–14. doi:10.1007/s11103-006-0026-7. PMID 16830182. S2CID 7840648.
- Seibt KM, Schmidt T, Heitkam T (February 2020). "The conserved 3' Angio-domain defines a superfamily of short interspersed nuclear elements (SINEs) in higher plants". The Plant Journal. 101 (3): 681–699. doi:10.1111/tpj.14567. PMID 31610059.
- Scarpato M, Angelini C, Cocca E, Pallotta MM, Morescalchi MA, Capriglione T (September 2015). "Short interspersed DNA elements and miRNAs: a novel hidden gene regulation layer in zebrafish?". Chromosome Research. 23 (3): 533–44. doi:10.1007/s10577-015-9484-6. PMID 26363800. S2CID 16759020.
- Ambros V (September 2004). "The functions of animal microRNAs". Nature. 431 (7006): 350–5. Bibcode:2004Natur.431..350A. doi:10.1038/nature02871. PMID 15372042. S2CID 205210153.
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (October 2004). "MicroRNA genes are transcribed by RNA polymerase II". The EMBO Journal. 23 (20): 4051–60. doi:10.1038/sj.emboj.7600385. PMC 524334. PMID 15372072.
- Faller M, Guo F (November 2008). "MicroRNA biogenesis: there's more than one way to skin a cat". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1779 (11): 663–7. doi:10.1016/j.bbagrm.2008.08.005. PMC 2633599. PMID 18778799.
- Lau NC, Lim LP, Weinstein EG, Bartel DP (October 2001). "An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans". Science. 294 (5543): 858–62. Bibcode:2001Sci...294..858L. doi:10.1126/science.1065062. PMID 11679671. S2CID 43262684.
- Cai X, Hagedorn CH, Cullen BR (December 2004). "Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs". RNA. 10 (12): 1957–66. doi:10.1261/rna.7135204. PMC 1370684. PMID 15525708.
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. (September 2003). "The nuclear RNase III Drosha initiates microRNA processing". Nature. 425 (6956): 415–9. Bibcode:2003Natur.425..415L. doi:10.1038/nature01957. PMID 14508493. S2CID 4421030.
- Bartel DP (January 2004). "MicroRNAs: genomics, biogenesis, mechanism, and function". Cell. 116 (2): 281–97. doi:10.1016/s0092-8674(04)00045-5. PMID 14744438.
- Schwarz DS, Zamore PD (May 2002). "Why do miRNAs live in the miRNP?". Genes & Development. 16 (9): 1025–31. doi:10.1101/gad.992502. PMID 12000786.
- Pratt AJ, MacRae IJ (July 2009). "The RNA-induced silencing complex: a versatile gene-silencing machine". The Journal of Biological Chemistry. 284 (27): 17897–901. doi:10.1074/jbc.R900012200. PMC 2709356. PMID 19342379.
- Nätt D, Johansson I, Faresjö T, Ludvigsson J, Thorsell A (2015). "High cortisol in 5-year-old children causes loss of DNA methylation in SINE retrotransposons: a possible role for ZNF263 in stress-related diseases". Clinical Epigenetics. 7 (1): 91. doi:10.1186/s13148-015-0123-z. PMC 4559301. PMID 26339299.
- Pal A, Srivastava T, Sharma MK, Mehndiratta M, Das P, Sinha S, Chattopadhyay P (November 2010). "Aberrant methylation and associated transcriptional mobilization of Alu elements contributes to genomic instability in hypoxia". Journal of Cellular and Molecular Medicine. 14 (11): 2646–54. doi:10.1111/j.1582-4934.2009.00792.x. PMC 4373486. PMID 19508390.
- Peterson M, Chandler VL, Bosco G (April 2013). "High SINE RNA Expression Correlates with Post-Transcriptional Downregulation of BRCA1". Genes. 4 (2): 226–43. doi:10.3390/genes4020226. PMC 3899967. PMID 24705161.
- Vanin EF (1985). "Processed pseudogenes: characteristics and evolution". Annual Review of Genetics. 19: 253–72. doi:10.1146/annurev.ge.19.120185.001345. PMID 3909943.
- Dewannieux M, Esnault C, Heidmann T (September 2003). "LINE-mediated retrotransposition of marked Alu sequences". Nature Genetics. 35 (1): 41–8. doi:10.1038/ng1223. PMID 12897783. S2CID 32151696.
- Jurka J (December 2004). "Evolutionary impact of human Alu repetitive elements". Current Opinion in Genetics & Development. 14 (6): 603–8. doi:10.1016/j.gde.2004.08.008. PMID 15531153.