Chymosin
Chymosin /ˈkaɪməsɪn/ or rennin /ˈrɛnɪn/ is a protease found in rennet. It is an aspartic endopeptidase belonging to MEROPS A1 family. It is produced by newborn ruminant animals in the lining of the abomasum to curdle the milk they ingest, allowing a longer residence in the bowels and better absorption. It is widely used in the production of cheese.
| Chymosin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
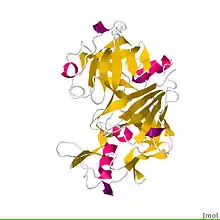 Crystal structure of bovine chymosin complex with the inhibitor CP-113972.[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 3.4.23.4 | ||||||||
| CAS no. | 9001-98-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Historically, chymosin was obtained by extracting it from the stomachs of slaughtered calves. Today, most commercial chymosin used in cheese production is produced recombinantly in E. coli, Aspergillus niger var awamori, and K. lactis.
Occurrence
Chymosin is found in a wide range of tetrapods,[2] although it is best known to be produced by ruminant animals in the lining of the abomasum. Chymosin is produced by gastric chief cells in newborn mammals[3] to curdle the milk they ingest, allowing a longer residence in the bowels and better absorption. Non-ruminant species that produce chymosin include pigs, cats, seals,[4] and chicks.[2]
One study reported finding a chymosin-like enzyme in some human infants,[5] but others have failed to replicate this finding.[6] Humans have a pseudogene for chymosin that does not generate a protein, found on chromosome 1.[4][7] Humans have other proteins to digest milk, such as pepsin and lipase.[8]: 262
In addition to the primate lineage leading up to humans, some other mammals have also lost the chymosin gene.[2]
Enzymatic reaction
Chymosin is used to bring about the extensive precipitation and curd formation in cheese-making. The native substrate of chymosin is K-casein which is specifically cleaved at the peptide bond between amino acid residues 105 and 106, phenylalanine and methionine.[9] The resultant product is calcium phosphocaseinate. When the specific linkage between the hydrophobic (para-casein) and hydrophilic (acidic glycopeptide) groups of casein is broken, the hydrophobic groups unite and form a 3D network that traps the aqueous phase of the milk.
Charge interactions between histidines on the kappa-casein and glutamates and aspartates of chymosin initiate enzyme binding to the substrate.[9] When chymosin is not binding substrate, a beta-hairpin, sometimes referred to as "the flap," can hydrogen bond with the active site, therefore covering it and not allowing further binding of substrate.[1]
Examples
Listed below are the ruminant Cym gene and corresponding human pseudogene:
|
| ||||||||||||||||||||||||||||||||||||||||||||
Recombinant chymosin
Because of the imperfections and scarcity of microbial and animal rennets, producers sought replacements. With the development of genetic engineering, it became possible to extract rennet-producing genes from animal stomach and insert them into certain bacteria, fungi or yeasts to make them produce chymosin during fermentation.[11][12] The genetically modified microorganism is killed after fermentation and chymosin is isolated from the fermentation broth, so that the fermentation-produced chymosin (FPC) used by cheese producers does not contain any GM component or ingredient.[13] FPC contains the identical chymosin as the animal source, but produced in a more efficient way. FPC products have been on the market since 1990 and are considered the ideal milk-clotting enzyme.[14]
FPC was the first artificially produced enzyme to be registered and allowed by the US Food and Drug Administration. In 1999, about 60% of US hard cheese was made with FPC[15] and it has up to 80% of the global market share for rennet.[16]
By 2008, approximately 80% to 90% of commercially made cheeses in the US and Britain were made using FPC.[13] The most widely used fermentation-produced chymosin is produced either using the fungus Aspergillus niger or using Kluyveromyces lactis.
FPC contains only chymosin B,[17] achieving a higher degree of purity compared with animal rennet. FPC can deliver several benefits to the cheese producer compared with animal or microbial rennet, such as higher production yield, better curd texture and reduced bitterness.[14]
References
- PDB: 1CZI; Groves MR, Dhanaraj V, Badasso M, Nugent P, Pitts JE, Hoover DJ, Blundell TL (October 1998). "A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure" (PDF). Protein Engineering. 11 (10): 833–40. doi:10.1093/protein/11.10.833. PMID 9862200.
- Lopes-Marques M, Ruivo R, Fonseca E, Teixeira A, Castro LF (November 2017). "Unusual loss of chymosin in mammalian lineages parallels neo-natal immune transfer strategies". Molecular Phylogenetics and Evolution. 116: 78–86. doi:10.1016/j.ympev.2017.08.014. PMID 28851538.
- Kitamura N, Tanimoto A, Hondo E, Andrén A, Cottrell DF, Sasaki M, Yamada J (August 2001). "Immunohistochemical study of the ontogeny of prochymosin--and pepsinogen-producing cells in the abomasum of sheep". Anatomia, Histologia, Embryologia. 30 (4): 231–5. doi:10.1046/j.1439-0264.2001.00326.x. PMID 11534329. S2CID 7552821.
- Staff, Online Mendelian Inheritance in Man (OMIM) Database. Last updated February 21, 1997 Chymosin pseudogene; CYMP prochymosin, included, in the OMIM
- Henschel MJ, Newport MJ, Parmar V (1987). "Gastric proteases in the human infant". Biology of the Neonate. 52 (5): 268–72. doi:10.1159/000242719. PMID 3118972.
- Szecsi PB, Harboe M (2013). "Chapter 5: Chymosin". In Rawlings ND, Salvesen G (eds.). Handbook of Proteolytic Enzymes. Vol. 1. pp. 37–42. doi:10.1016/B978-0-12-382219-2.00005-3.
- Fox PF (28 February 1999). Cheese: Chemistry, Physics and Microbiology. ISBN 9780834213388.
- Sanderson IR, Walker WA (1999). Development of the gastrointestinal tract. Hamilton, Ontario: B.C. Decker. ISBN 978-1-55009-081-9.
- Gilliland GL, Oliva MT, Dill J (1991). "Functional Implications of the Three-Dimensional Structure of Bovine Chymosin". Structure and Function of the Aspartic Proteinases. Advances in Experimental Medicine and Biology. Vol. 306. pp. 23–37. doi:10.1007/978-1-4684-6012-4_3. ISBN 978-1-4684-6014-8. PMID 1812710.
- PDB: 4CMS; Newman M, Safro M, Frazao C, Khan G, Zdanov A, Tickle IJ, et al. (October 1991). "X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin". Journal of Molecular Biology. 221 (4): 1295–309. doi:10.1016/0022-2836(91)90934-X. PMID 1942052.
- Emtage JS, Angal S, Doel MT, Harris TJ, Jenkins B, Lilley G, Lowe PA (June 1983). "Synthesis of calf prochymosin (prorennin) in Escherichia coli". Proceedings of the National Academy of Sciences of the United States of America. 80 (12): 3671–5. Bibcode:1983PNAS...80.3671E. doi:10.1073/pnas.80.12.3671. PMC 394112. PMID 6304731.
- Harris TJ, Lowe PA, Lyons A, Thomas PG, Eaton MA, Millican TA, et al. (April 1982). "Molecular cloning and nucleotide sequence of cDNA coding for calf preprochymosin". Nucleic Acids Research. 10 (7): 2177–87. doi:10.1093/nar/10.7.2177. PMC 320601. PMID 6283469.
- "Chymosin". GMO Compass. Archived from the original on 2015-03-26. Retrieved 2011-03-03.
- Law BA (2010). Technology of Cheesemaking. UK: Wiley-Blackwell. pp. 100–101. ISBN 978-1-4051-8298-0.
- "Food Biotechnology in the United States: Science, Regulation, and Issues". U.S. Department of State. Retrieved 2006-08-14.
- Johnson ME, Lucey JA (April 2006). "Major technological advances and trends in cheese". Journal of Dairy Science. 89 (4): 1174–8. doi:10.3168/jds.S0022-0302(06)72186-5. PMID 16537950.
- Bovine chymosins A and B differ by one amino acid residue. This is probably an alleic variant, according to Uniprot:P00794. The two isoforms have identical catalytic activity, so any improvement in the product is due to the elimination of other impurities.
Further reading
- Foltmann B (1966). "A review on prorennin and rennin". Comptes-Rendus des Travaux du Laboratoire Carlsberg. 35 (8): 143–231. PMID 5330666.
- Visser S, Slangen CJ, van Rooijen PJ (June 1987). "Peptide substrates for chymosin (rennin). Interaction sites in kappa-casein-related sequences located outside the (103-108)-hexapeptide region that fits into the enzyme's active-site cleft". The Biochemical Journal. 244 (3): 553–8. doi:10.1042/bj2440553. PMC 1148031. PMID 3128264.