Transferrin
Transferrins are glycoproteins found in vertebrates which bind and consequently mediate the transport of iron (Fe) through blood plasma.[5] They are produced in the liver and contain binding sites for two Fe3+ ions.[6] Human transferrin is encoded by the TF gene and produced as a 76 kDa glycoprotein.[7][8]
| Transferrin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Transferrin | ||||||||
| Pfam | PF00405 | ||||||||
| InterPro | IPR001156 | ||||||||
| PROSITE | PDOC00182 | ||||||||
| SCOP2 | 1lcf / SCOPe / SUPFAM | ||||||||
| |||||||||
Transferrin glycoproteins bind iron tightly, but reversibly. Although iron bound to transferrin is less than 0.1% (4 mg) of total body iron, it forms the most vital iron pool with the highest rate of turnover (25 mg/24 h). Transferrin has a molecular weight of around 80 kDa and contains two specific high-affinity Fe(III) binding sites. The affinity of transferrin for Fe(III) is extremely high (association constant is 1020 M−1 at pH 7.4)[9] but decreases progressively with decreasing pH below neutrality. Transferrins are not limited to only binding to iron but also to different metal ions.[10] These glycoproteins are located in various bodily fluids of vertebrates.[11][12] Some invertebrates have proteins that act like transferrin found in the hemolymph.[11][13]
When not bound to iron, transferrin is known as "apotransferrin" (see also apoprotein).
Occurrence and function
Transferrins are glycoproteins that are often found in biological fluids of vertebrates. When a transferrin protein loaded with iron encounters a transferrin receptor on the surface of a cell, e.g., erythroid precursors in the bone marrow, it binds to it and is transported into the cell in a vesicle by receptor-mediated endocytosis.[14] The pH of the vesicle is reduced by hydrogen ion pumps (H+
ATPases) to about 5.5, causing transferrin to release its iron ions.[11] Iron release rate is dependent on several factors including pH levels, interactions between lobes, temperature, salt, and chelator.[14] The receptor with its ligand bound transferrin is then transported through the endocytic cycle back to the cell surface, ready for another round of iron uptake.
Each transferrin molecule has the ability to carry two iron ions in the ferric form (Fe3+
).[13]
Humans and other mammals
The liver is the main site of transferrin synthesis but other tissues and organs, including the brain, also produce transferrin. A major source of transferrin secretion in the brain is the choroid plexus in the ventricular system.[15] The main role of transferrin is to deliver iron from absorption centers in the duodenum and white blood cell macrophages to all tissues. Transferrin plays a key role in areas where erythropoiesis and active cell division occur.[16] The receptor helps maintain iron homeostasis in the cells by controlling iron concentrations.[16]
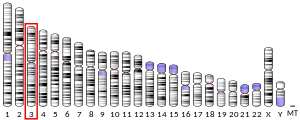
The gene coding for transferrin in humans is located in chromosome band 3q21.[7]
Medical professionals may check serum transferrin level in iron deficiency and in iron overload disorders such as hemochromatosis.
Other species
Drosophila melanogaster has three transferrin genes and is highly divergent from all other model clades, Ciona intestinalis one, Danio rerio has three highly divergent from each other, as do Takifugu rubripes and Xenopus tropicalis and Gallus gallus, while Monodelphis domestica has two divergent orthologs, and Mus musculus has two relatively close and one more distant ortholog. Relatedness and orthology/paralogy data are also available for Dictyostelium discoideum, Arabidopsis thaliana, and Pseudomonas aeruginosa.[17]
Structure
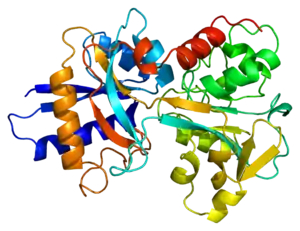
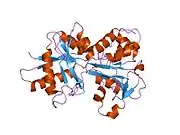
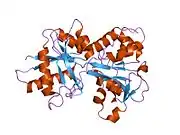
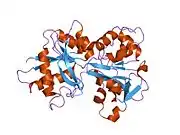
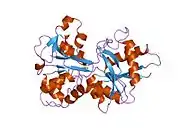
In humans, transferrin consists of a polypeptide chain containing 679 amino acids and two carbohydrate chains. The protein is composed of alpha helices and beta sheets that form two domains.[18] The N- and C- terminal sequences are represented by globular lobes and between the two lobes is an iron-binding site.[12]
The amino acids which bind the iron ion to the transferrin are identical for both lobes; two tyrosines, one histidine, and one aspartic acid. For the iron ion to bind, an anion is required, preferably carbonate (CO2−
3).[18][13]
Transferrin also has a transferrin iron-bound receptor; it is a disulfide-linked homodimer.[16] In humans, each monomer consists of 760 amino acids. It enables ligand bonding to the transferrin, as each monomer can bind to one or two atoms of iron. Each monomer consists of three domains: the protease, the helical, and the apical domains. The shape of a transferrin receptor resembles a butterfly based on the intersection of three clearly shaped domains.[18] Two main transferrin receptors found in humans denoted as transferrin receptor 1 (TfR1) and transferrin receptor 2 (TfR2). Although both are similar in structure, TfR1 can only bind specifically to human TF where TfR2 also has the capability to interact with bovine TF.[8]
Immune system
Transferrin is also associated with the innate immune system. It is found in the mucosa and binds iron, thus creating an environment low in free iron that impedes bacterial survival in a process called iron withholding. The level of transferrin decreases in inflammation.[21]
Role in disease
An increased plasma transferrin level is often seen in patients with iron deficiency anemia, during pregnancy, and with the use of oral contraceptives, reflecting an increase in transferrin protein expression. When plasma transferrin levels rise, there is a reciprocal decrease in percent transferrin iron saturation, and a corresponding increase in total iron binding capacity in iron deficient states[22]
A decreased plasma transferrin level can occur in iron overload diseases and protein malnutrition. An absence of transferrin results from a rare genetic disorder known as atransferrinemia, a condition characterized by anemia and hemosiderosis in the heart and liver that leads to heart failure and many other complications as well as to H63D syndrome.
Studies reveal that a transferrin saturation (serum iron concentration ÷ total iron binding capacity) over 60 percent in men and over 50 percent in women identified the presence of an abnormality in iron metabolism (Hereditary hemochromatosis, heterozygotes and homozygotes) with approximately 95 percent accuracy. This finding helps in the early diagnosis of Hereditary hemochromatosis, especially while serum ferritin still remains low. The retained iron in Hereditary hemochromatosis is primarily deposited in parenchymal cells, with reticuloendothelial cell accumulation occurring very late in the disease. This is in contrast to transfusional iron overload in which iron deposition occurs first in the reticuloendothelial cells and then in parenchymal cells. This explains why ferritin levels remain relative low in Hereditary hemochromatosis, while transferrin saturation is high.[23][24]
Transferrin and its receptor have been shown to diminish tumour cells when the receptor is used to attract antibodies.[16]
Transferrin and nanomedicine
Many drugs are hindered when providing treatment when crossing the blood-brain barrier yielding poor uptake into areas of the brain. Transferrin glycoproteins are able to bypass the blood-brain barrier via receptor-mediated transport for specific transferrin receptors found in the brain capillary endothelial cells.[25] Due to this functionality, it is theorized that nanoparticles acting as drug carriers bound to transferrin glycoproteins can penetrate the blood-brain barrier allowing these substances to reach the diseased cells in the brain.[26] Advances with transferrin conjugated nanoparticles can lead to non-invasive drug distribution in the brain with potential therapeutic consequences of central nervous system (CNS) targeted diseases (e.g. Alzheimer's or Parkinson's disease).[27]
Other effects
Carbohydrate deficient transferrin increases in the blood with heavy ethanol consumption and can be monitored through laboratory testing.[28]
Transferrin is an acute phase protein and is seen to decrease in inflammation, cancers, and certain diseases (in contrast to other acute phase proteins, e.g., C-reactive protein, which increase in case of acute inflammation).[29]
Pathology
Atransferrinemia is associated with a deficiency in transferrin.
In nephrotic syndrome, urinary loss of transferrin, along with other serum proteins such as thyroxine-binding globulin, gammaglobulin, and anti-thrombin III, can manifest as iron-resistant microcytic anemia.
Reference ranges
An example reference range for transferrin is 204–360 mg/dL.[30] Laboratory test results should always be interpreted using the reference range provided by the laboratory that performed the test.

A high transferrin level may indicate an iron deficiency anemia. Levels of serum iron and total iron binding capacity (TIBC) are used in conjunction with transferrin to specify any abnormality. See interpretation of TIBC. Low transferrin likely indicates malnutrition.
Interactions
Transferrin has been shown to interact with insulin-like growth factor 2[31] and IGFBP3.[32] Transcriptional regulation of transferrin is upregulated by retinoic acid.[33]
Related proteins
Members of the family include blood serotransferrin (or siderophilin, usually simply called transferrin); lactotransferrin (lactoferrin); milk transferrin; egg white ovotransferrin (conalbumin); and membrane-associated melanotransferrin.[34]
See also
References
- GRCh38: Ensembl release 89: ENSG00000091513 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000032554 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Crichton RR, Charloteaux-Wauters M (May 1987). "Iron transport and storage". European Journal of Biochemistry. 164 (3): 485–506. doi:10.1111/j.1432-1033.1987.tb11155.x. PMID 3032619.
- Hall DR, Hadden JM, Leonard GA, Bailey S, Neu M, Winn M, Lindley PF (January 2002). "The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively". Acta Crystallographica. Section D, Biological Crystallography. 58 (Pt 1): 70–80. doi:10.1107/s0907444901017309. PMID 11752780.
- Yang F, Lum JB, McGill JR, Moore CM, Naylor SL, van Bragt PH, et al. (May 1984). "Human transferrin: cDNA characterization and chromosomal localization". Proceedings of the National Academy of Sciences of the United States of America. 81 (9): 2752–6. Bibcode:1984PNAS...81.2752Y. doi:10.1073/pnas.81.9.2752. PMC 345148. PMID 6585826.
- Kawabata H (March 2019). "Transferrin and transferrin receptors update". Free Radical Biology & Medicine. 133: 46–54. doi:10.1016/j.freeradbiomed.2018.06.037. PMID 29969719. S2CID 49674402.
- Aisen P, Leibman A, Zweier J (March 1978). "Stoichiometric and site characteristics of the binding of iron to human transferrin". The Journal of Biological Chemistry. 253 (6): 1930–7. doi:10.1016/S0021-9258(19)62337-9. PMID 204636.
- Nicotra S, Sorio D, Filippi G, De Gioia L, Paterlini V, De Palo EF, et al. (November 2017). "Terbium chelation, a specific fluorescent tagging of human transferrin. Optimization of conditions in view of its application to the HPLC analysis of carbohydrate-deficient transferrin (CDT)". Analytical and Bioanalytical Chemistry. 409 (28): 6605–6612. doi:10.1007/s00216-017-0616-z. PMID 28971232. S2CID 13929228.
- MacGillivray RT, Moore SA, Chen J, Anderson BF, Baker H, Luo Y, et al. (June 1998). "Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release". Biochemistry. 37 (22): 7919–28. doi:10.1021/bi980355j. PMID 9609685.
- Dewan JC, Mikami B, Hirose M, Sacchettini JC (November 1993). "Structural evidence for a pH-sensitive dilysine trigger in the hen ovotransferrin N-lobe: implications for transferrin iron release". Biochemistry. 32 (45): 11963–8. doi:10.1021/bi00096a004. PMID 8218271.
- Baker EN, Lindley PF (August 1992). "New perspectives on the structure and function of transferrins". Journal of Inorganic Biochemistry. 47 (3–4): 147–60. doi:10.1016/0162-0134(92)84061-q. PMID 1431877.
- Halbrooks PJ, He QY, Briggs SK, Everse SJ, Smith VC, MacGillivray RT, Mason AB (April 2003). "Investigation of the mechanism of iron release from the C-lobe of human serum transferrin: mutational analysis of the role of a pH sensitive triad". Biochemistry. 42 (13): 3701–7. doi:10.1021/bi027071q. PMID 12667060.
- Moos T (November 2002). "Brain iron homeostasis". Danish Medical Bulletin. 49 (4): 279–301. PMID 12553165.
- Macedo MF, de Sousa M (March 2008). "Transferrin and the transferrin receptor: of magic bullets and other concerns". Inflammation & Allergy - Drug Targets. 7 (1): 41–52. doi:10.2174/187152808784165162. PMID 18473900.
- Gabaldón T, Koonin EV (May 2013). "Functional and evolutionary implications of gene orthology". Nature Reviews. Genetics. Nature Portfolio. 14 (5): 360–6. doi:10.1038/nrg3456. PMC 5877793. PMID 23552219.
- "Transferrin Structure". St. Edward's University. 2005-07-18. Archived from the original on 2012-12-11. Retrieved 2009-04-24.
- PDB: 1suv; Cheng Y, Zak O, Aisen P, Harrison SC, Walz T (Feb 2004). "Structure of the human transferrin receptor-transferrin complex". Cell. 116 (4): 565–76. doi:10.1016/S0092-8674(04)00130-8. PMID 14980223. S2CID 2981917.
- PDB: 2nsu; Hafenstein S, Palermo LM, Kostyuchenko VA, Xiao C, Morais MC, Nelson CD, Bowman VD, Battisti AJ, Chipman PR, Parrish CR, Rossmann MG (Apr 2007). "Asymmetric binding of transferrin receptor to parvovirus capsids". Proceedings of the National Academy of Sciences of the United States of America. 104 (16): 6585–9. Bibcode:2007PNAS..104.6585H. doi:10.1073/pnas.0701574104. PMC 1871829. PMID 17420467.
- Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O, Ledue TB, Craig WY (1999). "Reference distributions for the negative acute-phase serum proteins, albumin, transferrin and transthyretin: a practical, simple and clinically relevant approach in a large cohort". Journal of Clinical Laboratory Analysis. 13 (6): 273–9. doi:10.1002/(SICI)1098-2825(1999)13:6<273::AID-JCLA4>3.0.CO;2-X. PMC 6808097. PMID 10633294.
- Miller JL (July 2013). "Iron deficiency anemia: a common and curable disease". Cold Spring Harbor Perspectives in Medicine. 3 (7): a011866. doi:10.1101/cshperspect.a011866. PMC 3685880. PMID 23613366.
- Bacon BR, Adams PC, Kowdley KV, Powell LW, Tavill AS (July 2011). "Diagnosis and management of hemochromatosis: 2011 practice guideline by the American Association for the Study of Liver Diseases". Hepatology. Baltimore, Md. 54 (1): 328–43. doi:10.1002/hep.24330. PMC 3149125. PMID 21452290.
- "Hemochromatosis". guidelinecentral.com.
- Ghadiri M, Vasheghani-Farahani E, Atyabi F, Kobarfard F, Mohamadyar-Toupkanlou F, Hosseinkhani H (October 2017). "Transferrin-conjugated magnetic dextran-spermine nanoparticles for targeted drug transport across blood-brain barrier". Journal of Biomedical Materials Research Part A. 105 (10): 2851–2864. doi:10.1002/jbm.a.36145. PMID 28639394.
- Gaspar R (February 2013). "Nanoparticles: Pushed off target with proteins". Nature Nanotechnology. 8 (2): 79–80. Bibcode:2013NatNa...8...79G. doi:10.1038/nnano.2013.11. PMID 23380930.
- Li S, Peng Z, Dallman J, Baker J, Othman AM, Blackwelder PL, Leblanc RM (September 2016). "Crossing the blood-brain-barrier with transferrin conjugated carbon dots: A zebrafish model study". Colloids and Surfaces. B, Biointerfaces. 145: 251–256. doi:10.1016/j.colsurfb.2016.05.007. PMID 27187189.
- Sharpe PC (November 2001). "Biochemical detection and monitoring of alcohol abuse and abstinence". Annals of Clinical Biochemistry. 38 (Pt 6): 652–64. doi:10.1258/0004563011901064. PMID 11732647. S2CID 12203099.
- Jain S, Gautam V, Naseem S (January 2011). "Acute-phase proteins: As diagnostic tool". Journal of Pharmacy & Bioallied Sciences. 3 (1): 118–27. doi:10.4103/0975-7406.76489. PMC 3053509. PMID 21430962.
- "Normal Reference Range Table". Interactive Case Study Companion to Pathological Basis of Disease. The University of Texas Southwestern Medical Center at Dallas. Archived from the original on 2011-12-25. Retrieved 2008-10-25.
Kumar V, Hagler HK (1999). Interactive Case Study Companion to Robbins Pathologic Basis of Disease (6th Edition (CD-ROM for Windows & Macintosh, Individual) ed.). W B Saunders Co. ISBN 0-7216-8462-9. - Storch S, Kübler B, Höning S, Ackmann M, Zapf J, Blum W, Braulke T (December 2001). "Transferrin binds insulin-like growth factors and affects binding properties of insulin-like growth factor binding protein-3". FEBS Letters. 509 (3): 395–8. doi:10.1016/S0014-5793(01)03204-5. PMID 11749962. S2CID 22895295.
- Weinzimer SA, Gibson TB, Collett-Solberg PF, Khare A, Liu B, Cohen P (April 2001). "Transferrin is an insulin-like growth factor-binding protein-3 binding protein". The Journal of Clinical Endocrinology and Metabolism. 86 (4): 1806–13. doi:10.1210/jcem.86.4.7380. PMID 11297622.
- Hsu SL, Lin YF, Chou CK (April 1992). "Transcriptional regulation of transferrin and albumin genes by retinoic acid in human hepatoma cell line Hep3B". The Biochemical Journal. 283 ( Pt 2) (2): 611–5. doi:10.1042/bj2830611. PMC 1131079. PMID 1315521.
- Chung MC (October 1984). "Structure and function of transferrin". Biochemical Education. 12 (4): 146–154. doi:10.1016/0307-4412(84)90118-3.
Further reading
- Hershberger CL, Larson JL, Arnold B, Rosteck PR, Williams P, DeHoff B, et al. (December 1991). "A cloned gene for human transferrin". Annals of the New York Academy of Sciences. 646 (1): 140–54. Bibcode:1991NYASA.646..140H. doi:10.1111/j.1749-6632.1991.tb18573.x. PMID 1809186. S2CID 19519911.
- Bowman BH, Yang FM, Adrian GS (1989). Transferrin: evolution and genetic regulation of expression. Advances in Genetics. Vol. 25. pp. 1–38. doi:10.1016/S0065-2660(08)60457-5. ISBN 9780120176250. PMID 3057819.
- Parkkinen J, von Bonsdorff L, Ebeling F, Sahlstedt L (August 2002). "Function and therapeutic development of apotransferrin". Vox Sanguinis. 83 (Suppl 1): 321–6. doi:10.1111/j.1423-0410.2002.tb05327.x. PMID 12617162. S2CID 5876134.
External links
- Transferrin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P02787 (Serotransferrin) at the PDBe-KB.