Protein adsorption
Adsorption (not to be mistaken for absorption) is the accumulation and adhesion of molecules, atoms, ions, or larger particles to a surface, but without surface penetration occurring. The adsorption of larger biomolecules such as proteins is of high physiological relevance, and as such they adsorb with different mechanisms than their molecular or atomic analogs. Some of the major driving forces behind protein adsorption include: surface energy, intermolecular forces, hydrophobicity, and ionic or electrostatic interaction. By knowing how these factors affect protein adsorption, they can then be manipulated by machining, alloying, and other engineering techniques to select for the most optimal performance in biomedical or physiological applications.
Relevance
Many medical devices and products come into contact with the internal surfaces of the body, such as surgical tools and implants. When a non-native material enters the body, the first step of the immune response takes place and host extracellular matrix and plasma proteins aggregate to the material in attempts to contain, neutralize, or wall-off the injurious agent.[1] These proteins can facilitate the attachment of various cell types such as osteoblasts and fibroblasts that can encourage tissue repair.[2] Taking this a step further, implantable devices can be coated with a bioactive material to encourage adsorption of specific proteins, fibrous capsule formation, and wound healing. This would reduce the risk of implant rejection and accelerate recovery by selecting for the necessary proteins and cells necessary for endothelialization. After the formation of the endothelium, the body will no longer be exposed to the foreign material, and will stop the immune response.
Proteins such as collagen or fibrin often serve as scaffolds for cell adhesion and cell growth. This is an integral part to the structural integrity of cell sheets and their differentiation into more complex tissue and organ structures. The adhesion properties of proteins to non-biological surfaces greatly influences whether or not cells can indirectly attach to them via scaffolds. An implant like a hip-stem replacement necessitates integration with the host tissues, and protein adsorption facilitates this integration.
Surgical tools can be designed to be sterilized more easily so that proteins do not remain adsorbed to a surface, risking cross-contamination. Some diseases such as Creutzfeldt–Jakob disease and kuru (both related to mad cow disease) are caused by the transmission of prions, which are errant or improperly folded forms of a normally native protein. Surgical tools contaminated with prions require a special method of sterilization to completely eradicate all trace elements of the misfolded protein, as they are resistant to many of the normally used cleansing methods.
However, in some cases, protein adsorption to biomaterials can be an extremely unfavorable event. The adhesion of clotting factors may induce thrombosis, which may lead to stroke or other blockages.[3] Some devices are intended to interact with the internal body environment such as sensors or drug-delivery vehicles, and protein adsorption would hinder their effectiveness.
Fundamentals of Protein Adsorption
Proteins are biomolecules that are composed of amino acid subunits. Each amino acid has a side chain that gains or loses charge depending on the pH of the surrounding environment, as well as its own individual polar/nonpolar qualities.[4]
Charged regions can greatly contribute to how that protein interacts with other molecules and surfaces, as well as its own tertiary structure (protein folding). As a result of their hydrophilicity, charged amino acids tend to be located on the outside of proteins, where they are able to interact with surfaces.[5] It is the unique combination of amino acids that gives a protein its properties. In terms of surface chemistry, protein adsorption is a critical phenomenon that describes the aggregation of these molecules on the exterior of a material. The tendency for proteins to remain attached to a surface depends largely on the material properties such as surface energy, texture, and relative charge distribution. Larger proteins are more likely to adsorb and remain attached to a surface due to the higher number of contact sites between amino acids and the surface (Figure 1).

Energy of Protein Adsorption
The fundamental idea behind spontaneous protein adsorption is that adsorption occurs when more energy is released than gained according to Gibbs law of free energy.
This is seen in the equation:
where:
- ∆ads is net change of the parameters
- G is Gibbs free energy
- T is the temperature (SI unit: kelvin)
- S is the entropy (SI unit: joule per kelvin)
- H is the enthalpy (SI unit: joule)
In order for the protein adsorption to occur spontaneously, ∆adsG must be a negative number.
Vroman Effect
Proteins and other molecules are constantly in competition with one another over binding sites on a surface. The Vroman Effect, developed by Leo Vroman, postulates that small and abundant molecules will be the first to coat a surface. However, over time, molecules with higher affinity for that particular surface will replace them. This is often seen in materials that contact the blood where fibrinogen will bind to the surface first and over time will be replaced by kininogen.[6]
Rate of Adsorption
In order for proteins to adsorb, they must first come into contact with the surface through one or more of these major transport mechanisms: diffusion, thermal convection, bulk flow, or a combination thereof. When considering the transport of proteins, it is clear how concentration gradients, temperature, protein size and flow velocity will influence the arrival of proteins to a solid surface. Under conditions of low flow and minimal temperature gradients, the adsorption rate can be modeled after the diffusion rate equation.[5]
Diffusion Rate equation
where:
- D is the diffusion coefficient
- n is the surface concentration of protein
- Co is the bulk concentration of proteins
- t is time
A higher bulk concentration and/or higher diffusion coefficient (inversely proportional to molecular size) results in a larger number of molecules arriving at the surface. The consequential protein surface interactions result in high local concentrations of adsorbed protein, reaching concentrations of up to 1000 times higher than in the bulk solution.[5] However, the body is much more complex, containing flow and convective diffusion, and these must be considered in the rate of protein adsorption.
Flow in a thin channel
and
where:
- C is concentration
- D is the diffusion coefficient
- V is the velocity of flow
- x is the distance down the channel
- γ is the wall shear rate
- b is the height of the channel
This equation[5] is especially applicable to analyzing protein adsorption to biomedical devices in arteries, e.g. stents.
Forces and Interactions influencing protein adsorption
The four fundamental classes of forces and interaction in protein adsorption are: 1) ionic or electrostatic interaction, 2) hydrogen bonding, 3) hydrophobic interaction (largely entropically driven), and 4) interactions of charge-transfer or particle electron donor/acceptor type.[7]
Ionic or Electrostatic Interactions
The charge of proteins is determined by the pKa of its amino acid side chains, and the terminal amino acid and carboxylic acid. Proteins with isoelectric point (pI) above physiological conditions have a positive charge and proteins with pI below physiological conditions have a negative charge. The net charge of the protein, determined by the sum charge of its constituents, results in electrophoretic migration in a physiologic electric field. These effects are short-range because of the high di-electric constant of water, however, once the protein is close to a charged surface, electrostatic coupling becomes the dominant force.[8]
Hydrogen Bonding
Water has as much propensity to form hydrogen bonds as any group in a polypeptide. During a folding and association process, peptide and amino acid groups exchange hydrogen bonds with water. Thus, hydrogen bonding does not have a strong stabilizing effect on protein adsorption in an aqueous medium.[9]
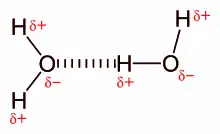 Illustration of two water molecules interacting to form a hydrogen bond
Illustration of two water molecules interacting to form a hydrogen bond
Hydrophobic Interactions
Hydrophobic interactions are essentially entropic interactions basically due to order/disorder phenomena in an aqueous medium. The free energy associated with minimizing interfacial areas is responsible for minimizing the surface area of water droplets and air bubbles in water. This same principle is the reason that hydrophobic amino acid side chains are oriented away from water, minimizing their interaction with water. The hydrophilic groups on the outside of the molecule result in protein water solubility. Characterizing this phenomenon can be done by treating these hydrophobic relationships with interfacial free energy concepts. Accordingly, one can think of the driving force of these interactions as the minimization of total interfacial free energy, i.e. minimization of surface area.[10]
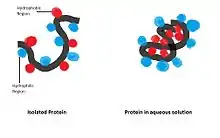
Charge-Transfer Interactions
Charge-transfer interactions are also important in protein stabilization and surface interaction. In general donor-acceptor processes, one can think of excess electron density being present which can be donated to an electrophilic species. In aqueous media, these solute interactions are primarily due to pi orbital electron effects.[11]
Other factors influencing protein adsorption
Temperature
Temperature has an effect on both, the equilibrium state and kinetics of protein adsorption. The amount of protein adsorbed at high temperature is usually higher than that at room temperature. Temperature variation causes conformational changes in protein influencing adsorption. These conformational rearrangements in proteins results in an entropy gain which acts as a major driving force for protein adsorption. The temperature effect on protein adsorption can be seen in food manufacturing processes, especially liquid foods such as, milk which causes severe fouling on the wall surfaces of equipment where thermal treatment is carried out.[12][13]
Ionic strength
Ionic strength determines the Debye length that correlates with the damping distance of the electric potential of a fixed charge in an electrolyte. So, higher the ionic strength the shorter are electrostatic interactions between charged entities. As a result, the adsorption of charged proteins to oppositely charged substrates is hindered whereas the adsorption to like charged substrates is enhanced, thereby influencing adsorption kinetics. Also, high ionic strength increases the tendency of proteins to aggregate.[12]
Multi-protein system
When a surface is exposed to a multi-protein solution, adsorption of certain protein molecules are favored over the others. Protein molecules approaching the surface compete for binding sites. In multi-protein system attraction between molecules can occur, whereas in single-protein solutions intermolecular repulsive interactions dominate. In addition, there is a time-dependent protein spreading, where protein molecules initially make contact with minimal binding sites on the surface. With the increase in protein's residence time on the surface, the protein may unfold for interaction with additional binding sites. This results in a time-dependent increase in the contact points between protein and surface. This further makes desorption less likely.[5]
Experimental approaches for studying protein adsorption
Solution depletion technique
This technique measures a concentration change of proteins in bulk solution before and after adsorption, Δcp. Any protein concentration change is attributed to the adsorbed layer, Γp.
Γp = Δcp V/Atot
where:
- V = total volume of protein solution
- Atot = Total area available for adsorption
This method also requires a high surface area material such as particulate and beaded adsorbents.[14]
Ellipsometry
Ellipsometry has been used widely for measuring protein adsorption kinetics as well as the structure of the adsorbed protein layer. It is an optical technique that measures the change of the polarization of light after reflection from a surface. This technique requires planar, reflecting surfaces, preferably quartz, silicon or silica, and a strong change in refractive index upon protein adsorption.[12]
Atomic-force microscopy
Atomic-force microscopy (AFM) is a powerful microscopy technique used for studying samples at a nanoscale and is often used to image protein distribution on a surface. It consists of a cantilever with a tip to scan over the surface. It is a valuable tool for measuring protein-protein and protein-surface interaction. However, the limiting factor of many AFM studies is that imaging is often performed after drying the surface which might affect protein folding and the structure of the protein layer. Moreover, the cantilever tip can dislodge a protein or corrugate the protein layer.[12][15]
Surface plasmon resonance
Surface plasmon resonance (SPR) has been widely used for measuring protein adsorption with high sensitivity. This technique is based on the excitation of surface plasmons, longitudinal electromagnetic waves originated at the interface between metals and dielectrics. The deposition on the conducting surface of molecules and thin layers within 200 nm modifies the dielectric properties of the system and thus the SPR response, signaling the presence of molecules on a metal surface.[16]
Quartz crystal microbalance
Quartz crystal microbalance (QCM) is an acoustic sensor built around a disk shaped quartz crystal. It makes use of the converse piezoelectric effect. QCM, and extended versions such as QCM-D, has been widely used for protein adsorption studies, especially, real time monitoring of label-free protein adsorption. In addition to the adsorption studies, QCM-D also provides information regarding elastic moduli, viscosity, and conformational changes [17]
Optical waveguide lightmode spectroscopy
Optical waveguide lightmode spectroscopy (OWLS) is a device that relies on a thin-film optical waveguide, enclosing a discrete number of guided electromagnetic waves. Guidance is achieved by means of a grating coupler. It is based on the measurements of effective refractive index of a thin-film layer above the waveguide. This technique works only on highly transparent surfaces.[17]
Other methods widely used for measuring the amount of protein adsorbed on surfaces include radio-labelling, Lowry assay, scanning angle reflectometry, total internal reflection fluorescence, bicinchoninic acid assay etc.
Protein Adsorption to Metals
Chemical composition
Metallic bonding refers to the specific bonding between positive metal ions and surrounding valence electron clouds.[18] This intermolecular force is relatively strong, and gives rise to the repeated crystalline orientation of atoms, also referred to as its lattice system. There are several types of common lattice formations, and each has its own unique packing density and atomic closeness. The negatively charged electron clouds of the metal ions will sterically hinder the adhesion of negatively charged protein regions due to charge repulsion, thus limiting the available binding sites of a protein to a metal surface.
The lattice formation can lead to connection with exposed potential metal-ion-dependent adhesion sites (MIDAS) which are binding sites for collagen and other proteins.[19] The surface of the metal has different properties than the bulk since the normal crystalline repeating subunits is terminated at the surface. This leaves the surface atoms without a neighboring atom on one side, which inherently alters the electron distribution. This phenomenon also explains why the surface atoms have a higher energy than the bulk, often simply referred to as surface energy. This state of higher energy is unfavorable, and the surface atoms will try to reduce it by binding to available reactive molecules.[20]
This is often accomplished by protein adsorption, where the surface atoms are reduced to a more advantageous energy state.
The internal environment of the body is often modeled to be an aqueous environment at 37 °C at pH 7.3 with plenty of dissolved oxygen, electrolytes, proteins, and cells.[5] When exposed to oxygen for an extended period of time, many metals may become oxidized and increase their surface oxidation state by losing electrons.[21] This new cationic state leaves the surface with a net positive charge, and a higher affinity for negatively charged protein side groups. Within the vast diversity of metals and metal alloys, many are susceptible to corrosion when implanted in the body. Elements that are more electronegative are corroded faster when exposed to an electrolyte-rich aqueous environment such as the human body.[22] Both oxidation and corrosion will lower the free energy, thus affecting protein adsorption as seen in Eq. 1.[23]
Effects of Topography
Surface roughness and texture has an undeniable influence on protein adsorption on all materials, but with the ubiquity of metal machining processes, it is useful to address how these impact protein behavior. The initial adsorption is important, as well as maintained adhesion and integrity. Research has shown that surface roughness can encourage the adhesion of scaffold proteins and osteoblasts, and results in an increase in surface mineralization.[24] Surfaces with more topographical features and roughness will have more exposed surface area for proteins to interact with.[5] In terms of biomedical engineering applications, micromachining techniques are often used to increase protein adhesion to implants in the hopes of shortening recovery time. The technique of laserpatterning introduces grooves and surface roughness that will influence adhesion, migration and alignment. Grit-blasting, a method analogous to sand blasting, and chemical etching have proven to be successful surface roughening techniques that promote the long-term stability of titanium implants.[25] The increase in stability is a direct result of the observed increase in extracellular matrix and collagen attachment, which results in increased osteoblast attachment and mineralization when compared to non-roughened surfaces.[26] Adsorption is not always desirable, however. Machinery can be negatively affected by adsorption, particularly with Protein adsorption in the food industry.
Protein adsorption to polymers[27]
Polymers are of great importance when considering protein adsorption in the biomedical arena. Polymers are composed of one or more types of "mers" bound together repeatedly, typically by directional covalent bonds. As the chain grows by the addition of mers, the chemical and physical properties of the material are dictated by the molecular structure of the monomer. By carefully selecting the type or types of mers in a polymer and its manufacturing process, the chemical and physical properties of a polymer can be highly tailored to adsorb specific proteins and cells for a particular application.
Conformation effects
Protein adsorption often results in significant conformational changes, which refers to changes in the secondary, tertiary, and quartary structures of proteins. In addition to adsorption rates and amounts, orientation and conformation are of critical importance. These conformational changes can affect protein interaction with ligands, substrates, and antigens which are dependent on the orientation of the binding site of interest. These conformational changes, as a result of protein adsorption, can also denature the protein and change its native properties.
Adsorption to polymer scaffolds
Tissue engineering is a relatively new field that utilizes a scaffolding as a platform upon which the desired cells proliferate. It is not clear what defines an ideal scaffold for a specific tissue type. The considerations are complex and protein adsorption only adds to the complexity. Although architecture, structural mechanics, and surface properties play a key role, understanding degradation and rate of protein adsorption are also key. In addition to the essentials of mechanics and geometry, a suitable scaffold construct will possess surface properties that are optimized for the attachment and migration of the cell types of particular interest.
Generally, it has been found that scaffolds that closely resemble the natural environments of the tissue being engineered are the most successful. As a result, much research has gone into investigating natural polymers that can be tailored, through processing methodology, toward specific design criteria. Chitosan is currently one of the most widely used polymers as it is very similar to naturally occurring glycosaminoglycan (GAGs) and it is degradable by human enzymes.[28]
Chitosan
Chitosan is a linear polysaccharide containing linked chitin-derived residues and is widely studied as a biomaterial due to its high compatibility with numerous proteins in the body. Chitosan is cationic and thus electrostatically reacts with numerous proteoglycans, anionic GAGs, and other molecules possessing a negative charge. Since many cytokines and growth factors are linked to GAG, scaffolds with the chitosan-GAG complexes are able to retain these proteins secreted by the adhered cells. Another quality of chitosan that gives it good biomaterial potential is its high charge density in solutions. This allows chitosan to form ionic complexes with many water-soluble anionic polymers, expanding the range of proteins that are able to bind to it and thus expanding its possible uses.[29]
| Polymer | Scaffold structure | Target tissue | Application cell type | Ref |
|---|---|---|---|---|
| Chitosan | 3D porous blocks | Bone | Osteoblast-like ROS | [30] |
| Chitosan-polyester | 3D fiber meshes | Bone | Human MSC | [31] |
| Chitosan-alginate | Injectable gel | Bone | Osteoblast-like MG63 | [32] |
| Chitosan-gelatin | 3D porous cylinders | Cartilage | Chondrocytes | [33] |
| Chitosan-GP | Injectable gel | Cartilage | Chondrocytes | [34] |
| Chitosan-collagen | Porous membranes | Skin | Fibroblast and keratinocyte co-culture | [35] |
Table 1: Structures, target tissues, and application cell types of chitosan-based scaffolds
}}
Protein adsorption prediction
Protein adsorption is critical for many industrial and biomedical applications. Accurate prediction of protein adsorption will enable progress to be made in these areas.
Biomolecular Adsorption Database
Biomolecular Adsorption Database (BAD) is a freely available online database with experimental protein adsorption data collected from the literature. The database can be used for the selection of materials for microfluidic device fabrication and for the selection of optimum operating conditions of lab-on-a-chip devices. The amount of protein adsorbed to the surface can be predicted using neural networks-based prediction available at BAD. This prediction has been validated to be below 5% error for the overall data available in the BAD. Other parameters, such as the thickness of protein layers and the surface tension of protein-covered surfaces, can also be estimated.[36]
References
- Rechendorff, Kristian. "The influence of surface roughness on protein adsorption" (PDF). Thesis. Interdisciplinary Nanoscience Center University of Aarhus, Denmark. Retrieved 23 May 2011.
- Maddikeri, RR; S. Tosatti; M. Schuler; S. Chessari; M. Textor; R.G. Richards; L.G. Harris (Feb 2008). "Reduced medical infection related bacterial strains adhesion on bioactive RGD modified titanium surfaces: A first step toward cell selective surfaces". Journal of Biomedical Materials Research Part A. 84A (2): 425–435. doi:10.1002/jbm.a.31323. PMID 17618480.
- Gorbet, MB; MV Sefton (Nov 2004). "Biomaterial-associated thrombosis: roles of coagulation factors, complement, platelets, and leukocytes". Biomaterials. 25 (26): 5681–5703. doi:10.1016/j.biomaterials.2004.01.023. PMID 15147815.
- Purdue. "Amino Acids". Retrieved 17 May 2011.
- Dee, Kay C (2002). An Introduction to Tissue-Biomaterial Interactions. Cal Poly Kennedy Library: John Wiley & Sons. pp. 1–50. ISBN 978-0-471-25394-5.
- Rosengren, Asa (2004). "Cell-protein-material Interactions on Bioceramics and Model Surfaces". Comprehensive Summaries of Uppsala Dissertations of the Faculty of Science and Technology.
- Ghosh, S; H.B. Bull (1966). "Adsorbed films of bovine serum albumin". Biochim. Biophys. Acta. 66: 150–157. doi:10.1016/0006-3002(63)91178-8. PMID 13947535.
- Andrade, Joseph D. (1985). Surface and interfacial Aspects of Biomedical Polymers. New York and London: Plenum. pp. 10–21. ISBN 978-0-306-41742-9.
- Cooper, A. (1980). "Conformational Fluctuations and Change in Biological Macromolecules". Scientific Progress. 66: 473–497.
- Tanford, C. (1981). The Hydrophobic Effect. New York: Wiley.
- Porath, J. (1979). "Charge-transfer Adsorption in Aqueous Media". Pure and Applied Chemistry. 51 (7): 1549–1559. doi:10.1351/pac197951071549. S2CID 38658475.
- Rabe, M. (2011). "Understanding protein adsorption at solid surfaces" (PDF). Advances in Colloid and Interface Science. 162 (1–2): 87–106. doi:10.1016/j.cis.2010.12.007. PMID 21295764.
- Nakanishi, K. (2001). "On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon". Journal of Bioscience and Bioengineering. 91 (3): 233–244. doi:10.1016/s1389-1723(01)80127-4. PMID 16232982.
- Hlady, V. (1999). Methods for Studying Protein Adsorption. pp. 402–429. doi:10.1016/S0076-6879(99)09028-X. ISBN 9780121822101. PMC 2664293. PMID 10507038.
{{cite book}}:|journal=ignored (help) - Lea, AS. (1992). "Manipulation of Proteins on Mica by Atomic Force Microscopy". Langmuir. 8 (1): 68–73. doi:10.1021/la00037a015. PMC 4137798. PMID 25147425.
- Servoli, E. (2009). "Comparative methods for the evaluation of protein adsorption". Macromolecular Bioscience. 9 (7): 661–670. doi:10.1002/mabi.200800301. hdl:10261/55283. PMID 19226562.
- Fulga, F.; D.V.Nicolau (2006). Biomolecular layers: Quantification of mass and thickness. Wiley Encyclopedia of Biomedical Engineering. doi:10.1002/9780471740360.ebs1351. ISBN 978-0-471-74036-0.
- Kopeliovich, Dimitri. "Metals Crystal Structure". SubsTech. Retrieved 17 May 2011.
- "The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding".
- Takeda, Satoshi; Makoto Fukawa; Yasuo Hayashi; Kiyoshi Matsumoto (8 Feb 1999). "Surface OH group governing adsorption properties of metal oxide films". Thin Solid Films. 339 (1–2): 220–224. Bibcode:1999TSF...339..220T. doi:10.1016/S0040-6090(98)01152-3.
- Over, H.; Seitsonen (20 September 2002). "A.P.". Science. 5589. 297 (5589): 2003–2005. doi:10.1126/science.1077063. PMID 12242427. S2CID 98612201.
- Xu, Liping; Guoning Yu; Erlin Zhang; Feng Pan; Ke Yang (4 June 2007). "In vivo corrosion behavior of Mg-Mn-Zn alloy for bone implant application". Journal of Biomedical Materials Research Part A. 83A (3): 703–711. doi:10.1002/jbm.a.31273. PMID 17549695.
- Park, Joon Bu (1984). Biomaterials Science and Engineering. Cal Poly Library: A Division of Plenum Publishing Corporation. pp. 171–181. ISBN 978-0-306-41689-7.
- Deligianni, DD; Katsala N; Ladas S; Sotiropoulou D; Amedee J; Missirlis YF (2001). "Effect of surface roughness of the titanium alloy Ti-6Al-4V on human bone marrow cell response and on protein adsorption". Biomaterials. 22 (11): 1241–1251. doi:10.1016/s0142-9612(00)00274-x. PMID 11336296.
- Hacking, SA; Harvey EJ; Tanzer M; Krygier JJ; Bobyn JD (2003). "Acid-etched microtexture for enhancement f bone growth into porous-coated implants". J Bone Joint Surg. 85B (8): 1182–1189. doi:10.1302/0301-620X.85B8.14233. PMID 14653605.
- Yang, SX; L Salvati; P Suh (23–25 September 2007). "How does silica grit-blasting affect Ti6Al4V alloy mineralization in a rat bone marrow cell culture system". Medical Device Materials. IV: 182–187.
- Firkowska-Boden, I.; Zhang, X.; Jandt, Klaus. D. (2017). "Controlling Protein Adsorption through Nanostructured Polymeric Surfaces". Advanced Healthcare Materials. 7 (1): 1700995. doi:10.1002/adhm.201700995. PMID 29193909. S2CID 35125391.
- Drury, J.L.; Mooney, D.J. (2003). "Hydrogels for tissue engineering: scaffold design variable and application". Biomaterials. 24 (24): 4337–4351. doi:10.1016/s0142-9612(03)00340-5. PMID 12922147.
- Van Blitterswijk, Clemens (2008). Tissue Engineering. Elsevier.
- Ho, Kuo; et al. (2004). "Preparation of porous scaffolds by using freeze-extraction and freeze-gelatin methods". Biomaterials. 25 (1): 129–138. doi:10.1016/s0142-9612(03)00483-6. PMID 14580916.
- Correlo, Vitor; Luciano F. Boesel; Mrinal Bhattacharya; Joao F. Mano; Nuno M. Neves; Ruis L. Reis (2005). "Hydroxyapatite Reinforced Chitosan and Polyester Blends for Biomedical Applications". Macromolecular Materials and Engineering. 290 (12): 1157–1165. doi:10.1002/mame.200500163. hdl:1822/13819.
- Li, Z; H. Ramay; K. Hauch; D. Xiao; M. Zhang (2005). "Chitosan-alginate hybrid scaffolds for bone tissue engineering". Biomaterials. 26 (18): 3919–3928. doi:10.1016/j.biomaterials.2004.09.062. PMID 15626439.
- Xia, W; Liu, W (2004). "Tissue engineering of cartilage with the use of chitosan-gelatin complex scaffolds". Journal of Biomedical Materials Research Part B: Applied Biomaterials. 71B (2): 373–380. doi:10.1002/jbm.b.30087. PMID 15386401.
- Chenite, A; C. Chaput; D. Wang; C. Combes; M.D. Buschmann; C.D. Hoemann; et al. (2000). "Novel injectable neutral solutions of chitosan form biodegradable gels in situ". Biomaterials. 21 (21): 2155–2161. doi:10.1016/s0142-9612(00)00116-2. PMID 10985488.
- Black, B; Bouez, C.; et al. (2005). "Optimization and characterization of an engineered human skin equivalent". Tissue Engineering. 11 (5–6): 723–733. doi:10.1089/ten.2005.11.723. PMID 15998214.
- Elena N Vasina; Ewa Paszek; Dan V Nicolau Jr; Dan V Nicolau (7 April 2009). "The BAD project: data mining, database and prediction of protein adsorption on surfaces". Lab Chip. 9 (7): 891–900. doi:10.1039/b813475h. PMID 19294299. Retrieved 31 October 2021.