OrthoFinder
OrthoFinder is a command-line software tool for comparative genomics.[1][2] OrthoFinder determines the correspondence between genes in different organisms (also known as orthology analysis). This correspondence provides a framework for understanding the evolution of life on Earth, and enables the extrapolation and transfer of biological knowledge between organisms.
| Original author(s) | David Emms Steven Kelly |
|---|---|
| Developer(s) | David Emms |
| Initial release | 2015 |
| Repository | github |
| Written in | Python |
| Operating system | Linux |
| Type | Bioinformatics |
| License | GNU GPL v3 |
| Website | github |
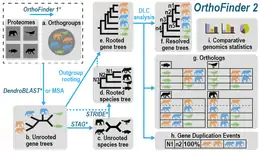
OrthoFinder workflow and outputs.
OrthoFinder takes FASTA files of protein sequences as input (one per species) and as output provides:
- Orthogroups
- Rooted Phylogenetic trees of all orthogroups
- A rooted species tree for the set of species included in the input dataset
- Hierarchical orthogroups for each node in the species tree
- Orthologs between all species
- Gene duplication events mapped to branches in the species tree
- Comparative genomic statistics
As of August 2021, the tool has been referenced by more than 1500 published studies.[3]
References
- Emms, David M; Kelly Steven (2015). "OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy". Genome Biology. 16 (157). doi:10.1186/s13059-015-0721-2. PMC 4531804. PMID 26243257.
- Emms, David M; Kelly Steven (2019). "OrthoFinder: phylogenetic orthology inference for comparative genomics". Genome Biology. 20 (238). doi:10.1186/s13059-019-1832-y. PMC 6857279. PMID 31727128.
- "Citations for Emms & Kelly 2015". Google Scholar. Retrieved 6 August 2021.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.