J. Richard McIntosh
J. Richard McIntosh is a Distinguished Professor Emeritus in Molecular, Cellular, and Developmental Biology at the University of Colorado Boulder.[1] McIntosh first graduated from Harvard with a BA in Physics in 1961, and again with a Ph.D. in Biophysics in 1968.[1] He began his teaching career at Harvard but has spent most of his career at the University of Colorado Boulder.[2] At the University of Colorado Boulder, McIntosh taught biology courses at both the undergraduate and graduate levels.[3] Additionally, he created an undergraduate course in the biology of cancer towards the last several years of his teaching career.[4] McIntosh's research career looks at a variety of things, including different parts of mitosis, microtubules, and motor proteins.
Research interests
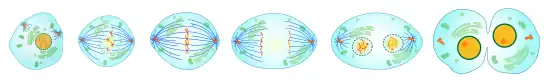
Mitosis
Most of McIntosh’s work focuses on the process of mitosis in the cell. Mitosis is the process of cell division that includes distinct movements of chromosomes in the cell and formation of mitotic spindles.[5] Additionally, McIntosh is very interested in the role of microtubules and motor proteins in this process. Mitotic spindles, composed of microtubules and other proteins, ensure that each of the two new cells during cell division both get one copy of every chromosome.[6] After the chromosomes are separated, then the cells are able to completely separate as well through cytokinesis.[6] In mitosis, there are multiple phases.[6] In prophase, the DNA starts to package itself for division and microtubules reorganize to prepare to form the mitotic spindle.[6] In prometaphase, kinetochores develop where the chromosome will attach to the mitotic spindle.[6] After that, chromosomes move towards the middle of the cell (metaphase plate) and the two copies separate during anaphase.[6] In some of McIntosh’s work he looks specifically at the anaphase A portion, which is related to where the chromosome is in relation to the pole it is moving towards.[6] Lastly, in telophase, the cell is wrapping up the stages of mitosis with creating a new nuclear envelope.[6]
Use of electron tomography
Additionally, in many of the works outlined below, McIntosh commonly uses electron tomography to image and study the cells. In electron tomography, many different images are put together to create a 3-D image of the subject being studied.[7] This technology is best suited to observe extremely complex structures and can image thinner sections of samples than can be physically made to study.[7]
1970s and 1980s
One of McIntosh’s earlier studies in the field of cell biology is in 1974, where his team published a paper on the structures of flagella of Pyrsonympha, an organism found in termites.[8] In this work, his team described the axostyle, a collection of microtubules, and presented that the axostyle’s attachment to other parts of the cell controls its function.[8]
In 1980, McIntosh’s curiosity with microtubules continued in “Visualization of the structural polarity of microtubules".[9] The polarity of microtubules is essential to generate the force needed to separate the chromosomes during mitosis, but at the time it was difficult to determine what the polarity is.[9] McIntosh’s team uses basal bodies and HeLa cells to study how protofilaments ‘hook’ onto them—either in right-handedness or left-handedness—in vitro to determine polarity.[9]
A few years later, McIntosh published a study in 1984 on how tubulin moves in mammalian cells with a focus on the cell cycle.[10] To study the movements in tubulin in cells during mitosis and interphase, McIntosh used two imaging methods: labeled (dichlorotriazinyl-aminofluorescein or DTAF-) tubulin and fluorescence redistribution (or recovery) after photobleaching (or FRAP).[10] Using the labeled tubulin, McIntosh observed how quickly the freely-added labeled tubulin was polymerized to the existing microtubule structures in the cell.[10] It was noted that measuring the tubulin addition in interphase was difficult due to the lack of structures, while it was more observable in a mitotic cell.[10] While using FRAP, McIntosh noticed that the tubulin redistributed throughout the cytoplasm in both a rapid phase as well as a slower phase.[10] Overall, the redistribution or movement of the labeled tubulin in cells undergoing mitosis was much faster than the redistribution observed for cells in interphase.[10] The next year, McIntosh’s interests started to shift towards motor proteins. Kinesin, a motor protein found to move around vesicles in the cell, was recently discovered on another paper published the same year.[11] Here, McIntosh explored the possibility of kinesin as an important part of mitosis, as it can be found in the mitotic spindle.[11] Some of the possible functions that McIntosh suggested kinesin may have in mitosis are that they move chromosomes down microtubules or move microtubules in different areas within the mitotic spindle.[11]
The next year, McIntosh’s interests started to shift towards motor proteins. Kinesin, a motor protein found to move around vesicles in the cell, was recently discovered on another paper published the same year.[11] Here, McIntosh explored the possibility of kinesin as an important part of mitosis, as it can be found in the mitotic spindle.[11] Some of the possible functions that McIntosh suggested kinesin may have in mitosis are that they move chromosomes down microtubules or move microtubules in different areas within the mitotic spindle.[11]
1990s
McIntosh’s interest in the connection of motor proteins and mitosis continues into the 1990s. In 1990, he published a paper on cytoplasmic dynein and mitosis.[12] Dynein, a protein first found in the flagella, had now been discovered in the cytoplasm as well.[12] It moves towards the minus end, or the slower-growing end, of the microtubules.[12] The research team used antibodies to image the distribution of cytoplasmic dynein in the cell.[12] Dynein was found to be near kinetochores during mitosis, while in interphase they were scattered around the cell.[12] The research team used this finding to suggest that dynein had a role in how chromosomes separate during mitosis due to the spatial differences of dynein during interphase and mitosis.[12]
In contrast to the topics McIntosh had researched so far, in 1996 his team published “Computer Visualization of Three-Dimensional Image Data using IMOD.” [13] In this article, his team shared the development of the computer software called IMOD.[13] With IMOD, researchers can study tomographies and data from both electron and light microscopes, and create three-dimensional images to interact with.[13] With these reconstructions, IMOD gives researchers the ability to help visualize their samples digitally.[7] The software is able to create collections of images and uses these collections to make models and measurements for further analysis.[7] One tool, the ‘slicer’, can help view samples at different angles.[7] Some other tools include zooming and panning (called the Zap window), a Model view window of a contour of the sample, an XYZ window that shows the other planes that cut through the point that is being studied, and the Tilt and Tumbler windows that show different projections that can be made.[13] The software can be downloaded and used for free at http://bio3d.colorado.edu.[7] This technology has been used to visualize cytoplasmic membranes, myofibrils, and the trans-Golgi network.[13]
A few years later in 1999, McIntosh’s team published a study using cryofixing and electron tomography to create a 3-dimensional model of the Golgi apparatus.[14] Two of the Golgi apparatus’s main functions are to modify proteins and to target them to their next destination.[14] For transport within the Golgi, McIntosh’s team proposed evidence that it can be done by vesicles in the cell that fuse with different membranes or by microtubules that are constantly forming and shifting around the Golgi.[14] By cryofixing, or rapidly freezing the cells to be studied, McIntosh’s team was able to preserve the cells essentially in time before using an electron microscope to visualize them.[14] In this work, the researchers visualized different coatings on the budding parts of the Golgi.[14] They were able to see the difference between clathrin-coated and non-clathrin-coated vesicles.[14] The buds help molecules in the Golgi be transported from the Golgi to their intended area in the cell.[14] In this work, the researchers describe the different cisternae, buds, vesicles, and coatings and the differences between the cis and trans faces of the Golgi.[14] This visualization technique led them to the conclusion that the different cisternae of the Golgi are not interconnected.[14] There are also many vesicles that surround the Golgi, most of which are not coated with clathrin.[14]
2000s
In 2002, McIntosh’s team continued his earlier interests in “Chromosome-microtubule interactions during mitosis.” [15] This review paper explains how spindle microtubules bind to chromosomes during segregation at places called kinetochores and the description of the Kinetochore-Dependent Checkpoint.[15] This point in the cell will block the segregation of chromosomes until all of the chromosomes are properly connected.[15] It also explains the role of microtubule proteins, motor proteins, and the microtubules themselves in the segregation process.[15] Some of the motor proteins mentioned in the article include those associated with the kinetochore at the plus or minus end or those that help with the disassembly of microtubules.[15] Some motor proteins associate with the chromosomes instead of the kinetochores, including chromokinesins, and some work at the spindle poles.[15]
McIntosh also contributed to a work published in 2004 by many scientists in “A standard kinesin nomenclature”.[16] In this work, these scientists helped create a naming structure for kinesin proteins, that are involved in transport in the cell along with microtubules.[16] The kinesin proteins were split into fourteen different families, and given different names such as kinesin-1, kinesin-2, etc.[16] This new structure of naming was created so that the existing kinesins can be easily classified as well as classify new kinesin proteins when they are discovered.[16] The authors recommend using protein sequence alignment based homology search to help classify the kinesins.[16] It also makes specifications on what makes a kinesin recognized.[16] Additionally, this paper also gives guidance on how to address kinesins (and their former names, if applicable) in papers.[16]
In 2005, McIntosh and his team published a study that investigated the link between the depolymerization of microtubules and the generation of force, showing how the chemistry of the tubulin dimers can create mechanical force.[17] The researchers attached glass microbeads to tubulin.[17] By using laser tweezers, McIntosh’s team was able to observe that the bead would move, typically towards the minus end, as the microtubules depolymerized off of it.[17] In particular, the bending of the protofilaments would cause the tweezers to pick up the force generated.[17] Here, the researchers applied these findings to chromosome segregation and concluded that the microtubule dynamics create the forces needed during mitosis.[17]
In 2006, McIntosh’s team used cyroelecton tomography to image axonemes in sea urchin sperm and Chlamydomonas reinhardtii.[18] Axonemes are structures in cilia and flagella consisting of a specific pattern of microtubules.[18] McIntosh’s team was particularly interested in the dynein protein.[18] In this paper, the researchers describe how the different subunits of dynein attach to the microtubules and how they might generate force as well.[18]
2010s
McIntosh’s letter “Motors or dynamics: What really moves chromosomes?” to Nature cell biology in 2012 explains an overview of the different directions his lab has taken so far.[19] In those who study how chromosomes move during the process of mitosis, researchers argue either that microtubules generate the forces needed to separate them or that certain motor proteins do.[19] When McIntosh first started his research, he noted he was very confident in the motor protein school of thought and related it to the sliding filament theory used to explain muscle contraction.[19] However, by reading other research group’s papers and conducting other studies that found adenosine triphosphate (ATP) and the motor protein dynein were not necessary for chromosome movement, he started to consider the influence that microtubules had in this process.[19]
Continuing on the interest of microtubules, the mitotic spindle, and mitosis, in 2013 his team published a paper studying the speed of chromosome movement during mitosis.[20] It was observed overall that microtubule depolymerization and the movement of motor proteins are very fast, but the chromosomes move slowly during their separation in the anaphase A stage of mitosis.[20] In this study, McIntosh and his team suggest that motor proteins could have an influence in the rate of movement by influencing depolymerization.[20] It was also noted that separation of different protofilaments over individual tubulin dimers may be another influence in the depolymerization and subsequent chromosome movement in mitosis.[20]
Focusing now on microtubule polymerization instead of depolymerization, in 2018 McIntosh’s team used electron tomography to study the microtubules in vitro as well as in six different species.[21] The researchers noticed as the microtubules grow, they bend out from the axis, similar to how the microtubules bend off when they are depolymerizing as well.[21]
2020s
Continuing using the electron tomography technique, in 2020 McIntosh and his team used this visualization strategy to observe how different kinds of microtubules work together during the metaphase state of mitosis.[22] One important structure in this phase is kinetochore microtubules or referred to as KMTs.[22] They, along with other structures in the cell, work together to balance out the different forces in the cell during mitosis.[22]
Books
Throughout his career, McIntosh has helped edit and write books as well. In 2001, he was an editor along with Joseph G. Gall (from the Carnegie Institution of Washington) in Landmark Papers in Cell Biology.[23] This work, celebrating the 40th anniversary of the creation of the American Society for Cell Biology (founded in 1960) includes 42 major papers in the field of cell biology along with the editor’s own thoughts.[23] Some of the topics in this book include transcription, mitosis, cell membrane, and the cytoskeleton.[23]
In 2017, McIntosh is the guest editor of Mechanisms of Mitotic Chromosome Segregation.[24] In this work, McIntosh connects various findings about mitosis from a variety of organisms.[24] In the introduction, McIntosh also recognizes the importance of how the evolution of our understanding of mitosis comes from the advancement of other technologies in the field such as better cameras, better ways to purify molecules, and an increase in the understanding of genetics.[24] This collection of review articles helps readers get an overview of mitosis, a process that McIntosh believes is essential to life itself.[24]
Published in 2019, Understanding Cancer: An Introduction to the Biology, Medicine, and Societal Implications of the Disease is a resource for those who want to learn holistically more about cancer.[4] This book, influenced by his son’s death to lung cancer, discusses every stage of the process: screening, diagnosing, and treating.[4] The text is written so that those without previous knowledge are able to learn the basics about cancer.[4] McIntosh recognizes in the introduction that cancer is just more than science; it encompasses other fields such as history and religion.[2] In this text, McIntosh discusses science-heavy topics related to cancer such as the role of oncogenes, tumor suppressors, and the immune system.[4] Additionally, the text covers topics beyond the scientific descriptions of cancer such as on the future of cancer, minimizing the risk of cancer, and living with cancer.[4]
Online lectures
Recorded in late 2008, McIntosh is featured on the website “iBiology” giving a series of talks titled “Eukaryotic cell division.” [2] In this series, the process is split up into three different videos specializing in chromosome division, experimentation, and the mitotic stage of anaphase A.[2]
Accolades
From 1984 to 2006, McIntosh served as the Director of the Boulder Laboratory for 3-D Electron Microscopy of Cells.[1] In 1994, McIntosh served as a Research Professor of the American Cancer Society and continued this role until 2006.[1] In 1994, McIntosh was also the President of the American Society for Cell Biology.[1] In 1999, McIntosh was elected to both the National Academy of Sciences and the American Academy of Arts and Sciences.[1] The National Academy of Sciences lists him as a primary member of the Cellular and Developmental Biology section and a secondary member of the Biophysics and Computational Biology section.[25] In 2000, McIntosh was award with the Distinguished Professor title at the University of Colorado, Boulder.[1] After he retired in 2006, McIntosh continues to conduct research and publish books and articles.[23]
References
- "J. Richard McIntosh". Biophysics Group. 2015-11-13. Retrieved 2021-04-15.
- "Richard 'Dick' McIntosh • iBiology". iBiology. Retrieved 2021-04-15.
- "Richard McIntosh - Routledge & CRC Press Author Profile". www.routledge.com. Retrieved 2021-04-15.
- McIntosh JR (2019). Understanding cancer : an introduction to the biology, medicine, and societal implications of this disease. Boca Raton. ISBN 978-0-8153-4535-0. OCLC 1066186470.
{{cite book}}: CS1 maint: location missing publisher (link) - McIntosh JR, Hays T (December 2016). "A Brief History of Research on Mitotic Mechanisms". Biology. 5 (4): 55. doi:10.3390/biology5040055. PMC 5192435. PMID 28009830.
- McIntosh JR (September 2016). "Mitosis". Cold Spring Harbor Perspectives in Biology. 8 (9): a023218. doi:10.1101/cshperspect.a023218. PMC 5008068. PMID 27587616.
- O'Toole ET, Winey M, McIntosh JR, Mastronarde DN (2002-01-01). "Electron tomography of yeast cells". Guide to Yeast Genetics and Molecular and Cell Biology Part C. Methods in Enzymology. Vol. 351. pp. 81–95. doi:10.1016/S0076-6879(02)51842-5. ISBN 9780121822545. PMC 4440791. PMID 12073377.
- Bloodgood RA, Miller KR, Fitzharris TP, Mcintosh JR (May 1974). "The ultrastructure of Pyrsonympha and its associated microorganisms". Journal of Morphology. 143 (1): 77–105. doi:10.1002/jmor.1051430104. PMID 30326674. S2CID 53528610.
- Heidemann SR, McIntosh JR (July 1980). "Visualization of the structural polarity of microtubules". Nature. 286 (5772): 517–9. Bibcode:1980Natur.286..517H. doi:10.1038/286517a0. PMID 7402333. S2CID 1656543.
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR (December 1984). "Tubulin dynamics in cultured mammalian cells". The Journal of Cell Biology. 99 (6): 2175–86. doi:10.1083/jcb.99.6.2175. PMC 2113582. PMID 6501419.
- Scholey JM, Porter ME, Grissom PM, McIntosh JR (December 1985). "Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle". Nature. 318 (6045): 483–6. Bibcode:1985Natur.318..483S. doi:10.1038/318483a0. PMID 2933590. S2CID 4345279.
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR (May 1990). "Cytoplasmic dynein is localized to kinetochores during mitosis". Nature. 345 (6272): 263–5. Bibcode:1990Natur.345..263P. doi:10.1038/345263a0. PMID 2139717. S2CID 4364671.
- Kremer JR, Mastronarde DN, McIntosh JR (1996-01-01). "Computer visualization of three-dimensional image data using IMOD". Journal of Structural Biology. 116 (1): 71–6. doi:10.1006/jsbi.1996.0013. PMID 8742726. S2CID 4608130.
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA (March 1999). "Golgi structure in three dimensions: functional insights from the normal rat kidney cell". The Journal of Cell Biology. 144 (6): 1135–49. doi:10.1083/jcb.144.6.1135. PMC 2150572. PMID 10087259.
- McIntosh JR, Grishchuk EL, West RR (2002-11-01). "Chromosome-microtubule interactions during mitosis". Annual Review of Cell and Developmental Biology. 18 (1): 193–219. doi:10.1146/annurev.cellbio.18.032002.132412. PMID 12142285.
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, et al. (October 2004). "A standardized kinesin nomenclature". The Journal of Cell Biology. 167 (1): 19–22. doi:10.1083/jcb.200408113. PMC 2041940. PMID 15479732.
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR (November 2005). "Force production by disassembling microtubules". Nature. 438 (7066): 384–8. Bibcode:2005Natur.438..384G. doi:10.1038/nature04132. PMID 16292315. S2CID 4415883.
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR (August 2006). "The molecular architecture of axonemes revealed by cryoelectron tomography". Science. 313 (5789): 944–8. Bibcode:2006Sci...313..944N. doi:10.1126/science.1128618. PMID 16917055. S2CID 43436284.
- McIntosh JR (December 2012). "Motors or dynamics: what really moves chromosomes?". Nature Cell Biology. 14 (12): 1234. doi:10.1038/ncb2649. PMC 4429872. PMID 23196840.
- Betterton MD, McIntosh JR (December 2013). "Regulation of chromosome speeds in mitosis". Cellular and Molecular Bioengineering. 6 (4): 418–430. doi:10.1007/s12195-013-0297-4. PMC 4578309. PMID 26405462.
- McIntosh JR, O'Toole E, Morgan G, Austin J, Ulyanov E, Ataullakhanov F, Gudimchuk N (August 2018). "Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments". The Journal of Cell Biology. 217 (8): 2691–2708. doi:10.1083/jcb.201802138. PMC 6080942. PMID 29794031.
- O'Toole E, Morphew M, McIntosh JR (February 2020). "Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase". Molecular Biology of the Cell. 31 (3): 184–195. doi:10.1091/mbc.E19-07-0405. PMC 7001478. PMID 31825721.
- Gall JG (November 2000). Landmark Papers in Cell Biology : Selected Research Articles Celebrating Forty Years of The ASCB. J. Richard McIntosh. Plymouth: Cold Spring Harbor Laboratory Press. ISBN 0-87969-602-8. OCLC 697711820.
- Mechanisms of mitotic chromosome segregation. [Place of publication not identified]: MDPI AG. 2017. ISBN 978-3-03842-402-4. OCLC 990847914.
- "J. Richard McIntosh". www.nasonline.org. Retrieved 2021-04-15.