Fructose 2,6-bisphosphate
Fructose 2,6-bisphosphate, abbreviated Fru-2,6-P2, is a metabolite that allosterically affects the activity of the enzymes phosphofructokinase 1 (PFK-1) and fructose 1,6-bisphosphatase (FBPase-1) to regulate glycolysis and gluconeogenesis. [1] Fru-2,6-P2 itself is synthesized and broken down by the bifunctional enzyme phosphofructokinase 2/fructose-2,6-bisphosphatase (PFK-2/FBPase-2).[2]
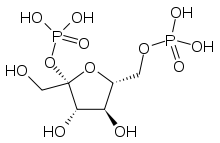 | |
| Names | |
|---|---|
| IUPAC name
2,6-Di-O-phosphono-β-D-fructofuranose | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
| MeSH | fructose+2,6-bisphosphate |
PubChem CID |
|
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C6H14O12P2 | |
| Molar mass | 340.114 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
The synthesis of Fru-2,6-P2 is performed through a bifunctional enzyme containing both PFK-2 and FBPase-2, which is dephosphorylated, allowing the PFK-2 portion to phosphorylate fructose 6-phosphate using ATP. The breakdown of Fru-2,6-P2 is catalyzed by the phosphorylation of the bifunctional enzyme, which allows FBPase-2 to dephosphorylate fructose 2,6-bisphosphate to produce fructose 6-phosphate and Pi.[3]
Effects on glucose metabolism
Fru-2,6-P2 strongly activates glucose breakdown in glycolysis through allosteric modulation (activation) of phosphofructokinase 1 (PFK-1). Elevated expression of Fru-2,6-P2 levels in the liver allosterically activates phosphofructokinase 1 by increasing the enzyme’s affinity for fructose 6-phosphate, while decreasing its affinity for inhibitory ATP and citrate. At physiological concentration, PFK-1 is almost completely inactive, but interaction with Fru-2,6-P2 activates the enzyme to stimulate glycolysis and enhance breakdown of glucose.[4]
Cellular stress as a result of oncogenesis or DNA damage among others, activates certain genes by the tumor suppressor p53. One such gene is for the expression of TP53-induced glycolysis and apoptosis regulator (TIGAR); an enzyme that inhibits glycolysis, monitors the cellular levels of reactive oxygen species, and protects cells from apoptosis. The structure of TIGAR is shown to be nearly identical to FBPase-2 on the bifunctional enzyme. TIGAR removes the allosteric effector, Fru-2,6-P2., therefore the activator does not enhance the affinity of the enzyme (PFK1) for its substrate (fructose 6-phosphate). Furthermore, TIGAR also removes the glycolytic intermediate fructose 1,6-bisphosphate, the product of the PFK catalyzed third reaction of glycolysis and the substrate for the following aldolase fourth reaction of glycolysis. [5]
Production regulation
The concentration of Fru-2,6-P2 in cells is controlled through regulation of the synthesis and breakdown by PFK-2/FBPase-2. The primary regulators of this are the hormones insulin, glucagon, and epinephrine which affect the enzyme through phosphorylation/dephosphorylation reactions. Release of the hormone glucagon triggers production of cyclic adenosine monophosphate (cAMP), which activates a cAMP-dependent protein kinase. This kinase phosphorylates the PFK-2/FBPase-2 enzyme at an NH2-terminal Ser residue with ATP to activate the FBPase-2 activity and inhibit the PFK-2 activity of the enzyme, thus reducing levels of Fru-2,6-P2 in the cell. With decreasing amounts of Fru-2,6-P2, glycolysis becomes inhibited while gluconeogenesis is activated. Insulin triggers the opposite response. As a phosphoprotein phosphatase, insulin dephosphorylates the enzyme, thus activating the PFK-2 and inhibiting the FBPase-2 activities. With additional Fru-2,6-P2 present, activation of PFK-1 occurs to stimulate glycolysis while inhibiting gluconeogenesis.[3][6]
Regulation of sucrose production
Fru-2,6-P2 plays an important role in the regulation of triose phosphates, the end products of the Calvin Cycle. In the Calvin Cycle, 5/6th of triose phosphates are recycled to make ribulose 1,5-bisphosphate. The remaining 1/6 of triose phosphate can be converted into sucrose or stored as starch. Fru-2,6-P2 inhibits production of fructose 6-phosphate, a necessary element for sucrose synthesis. When the rate of photosynthesis in the light reactions is high, triose phosphates are constantly produced and the production of Fru-2,6-P2 is inhibited, thus producing sucrose. Fru-2,6-P2 production is activated when plants are in the dark and photosynthesis and triose phosphates are not produced.[7]
References
- Alfarouk, Khalid O.; Verduzco, Daniel; Rauch, Cyril; Muddathir, Abdel Khalig; Bashir, Adil H. H.; Elhassan, Gamal O.; Ibrahim, Muntaser E.; Orozco, Julian David Polo; Cardone, Rosa Angela; Reshkin, Stephan J.; Harguindey, Salvador (18 December 2014). "Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question". Oncoscience. 1 (12): 777–802. doi:10.18632/oncoscience.109. PMC 4303887. PMID 25621294.
- Wu C, Khan SA, Peng LJ, Lange AJ (2006). "Roles for fructose-2,6-bisphosphate in the control of fuel metabolism: beyond its allosteric effects on glycolytic and gluconeogenic enzymes". Adv. Enzyme Regul. 46 (1): 72–88. doi:10.1016/j.advenzreg.2006.01.010. PMID 16860376.
- Kurland IJ, Pilkis SJ (June 1995). "Covalent control of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: insights into autoregulation of a bifunctional enzyme". Protein Sci. 4 (6): 1023–37. doi:10.1002/pro.5560040601. PMC 2143155. PMID 7549867.
- Lange AJ. "fructose-2,6-bisphosphate". University of Minnesota. Archived from the original on 2010-06-12.
- Garret, Reginald H.; Grisham, Charles M. (2013). Biochemistry. Belmont, CA: Brooks/Cole Cengage Learning. p. 730. ISBN 978-1-133-10629-6.
- Smith WE, Langer S, Wu C, Baltrusch S, Okar DA (June 2007). "Molecular coordination of hepatic glucose metabolism by the 6-phosphofructo-2-kinase/fructose-2,6- bisphosphatase:glucokinase complex". Mol. Endocrinol. 21 (6): 1478–87. doi:10.1210/me.2006-0356. PMID 17374851.
- Nielsen TH, Rung JH, Villadsen D (November 2004). "Fructose-2,6-bisphosphate: a traffic signal in plant metabolism". Trends Plant Sci. 9 (11): 556–63. doi:10.1016/j.tplants.2004.09.004. PMID 15501181.