Floxing
In genetics, floxing refers to the sandwiching of a DNA sequence (which is then said to be floxed) between two lox P sites. The terms are constructed upon the phrase "flanking/flanked by LoxP". Recombination between LoxP sites is catalysed by Cre recombinase. Floxing a gene allows it to be deleted (knocked out), translocated or inverted in a process called Cre-Lox recombination. [1] The floxing of genes is essential in the development of scientific model systems as it allows researchers to have spatial and temporal alteration of gene expression.[2] Moreover, animals such as mice can be used as models to study human disease. Therefore, Cre-lox system can be used in mice to manipulate gene expression in order to study human diseases and drug development.[3] For example, using the Cre-lox system, researchers can study oncogenes and tumor suppressor genes and their role in development and progression of cancer in mice models.[4]
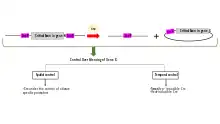
Uses in research
Floxing a gene allows it to be deleted (knocked out),[5][6] translocated or inserted[7] (through various mechanisms in Cre-Lox recombination).
The floxing of genes is essential in the development of scientific model systems as it allows spatial and temporal alteration of gene expression. In layman's terms, the gene can be knocked-out (inactivated) in a specific tissue in vivo, at any time chosen by the scientist. The scientist can then evaluate the effects of the knocked-out gene and identify the gene’s normal function[8]. This is different from having the gene absent starting from conception, whereby inactivation or loss of genes that are essential for the development of the organism may interfere with the normal function of cells and prevent the production of viable offspring.[9]
Mechanism of deletion
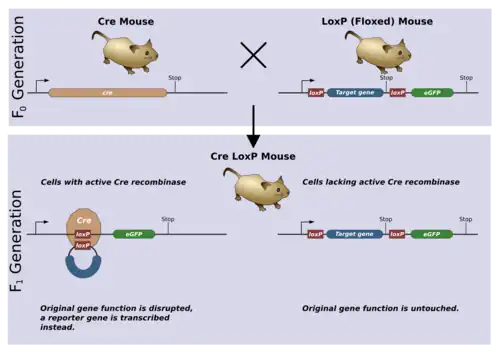
Deletion events are useful for performing gene editing experiments through precisely editing out segments of or even whole genes. Deletion requires floxing of the segment of interest with loxP sites which face the same direction. The Cre recombinase will detect the unidirectional loxP sites and excise the floxed segment of DNA.[10] The successfully edited clones can be selected using a selection marker which can be removed using the same Cre-loxP system.[10] The same mechanism can be used to create conditional alleles by introducing an FRT/Flp site which accomplishes the same mechanism but with a different enzyme.
Mechanism of inversion
Inversion events are useful for maintaining the amount of genetic material. The inverted genes are not often associated with abnormal phenotypes, meaning the inverted genes are generally viable.[11] Cre-loxP recombination that result in insertion requires loxP sites to flox the gene of interest, with the loxP sites oriented towards each other. By undergoing Cre recombination, the region floxed by the loxP sites will become inverted,[12] this process is not permanent and can be reversed.[13]
Mechanism of translocation
Translocation events occur when the loxP sites flox genes on two different DNA molecules in a unidirectional orientation. Cre recombinase is then used to generate a translocation between the two DNA molecules, exchanging the genetic material from one DNA molecule to the other forming a simultaneous translocation of both floxed genes.[14][15]
Common applications in research
Cardiomyocytes (heart muscle tissue) have been shown to express a type of Cre recombinase that is highly specific to cardiomyocytes and can be used by researchers to perform highly efficient recombinations. This is achieved by using a type of Cre whose expression is driven by the -myosin heavy chain promoter (-MyHC). These recombinations are capable of disrupting genes in a manner that is specific to only heart tissue in vivo and allows for the creation of conditional knockouts of the heart mostly for use as controls.[16]
For example, using the Cre recombinase with the -MyHC promoter causes the floxed gene to be inactivated in the heart alone. Further, these knockouts can be inducible. In several mouse studies, tamoxifen is used to induce the Cre recombinase.[17][18] In this case, Cre recombinase is fused to a portion of the mouse estrogen receptor (ER) which contains a mutation within its ligand binding domain (LBD). The mutation renders the receptor inactive, which leads to incorrect localization through its interactions with chaperone proteins such as heat shock protein 70 and 90 (Hsp70 and Hsp90). Tamoxifen binds to Cre-ER and disrupts its interactions with the chaperones, which allows the Cre-ER fusion protein to enter the nucleus and perform recombination on the floxed gene.[19][20] Additionally, Cre recombinase can be induced by heat when under the control of specific heat shock elements (HSEs).[21][22]
References
- Nagy A (February 2000). "Cre recombinase: the universal reagent for genome tailoring". Genesis. 26 (2): 99–109. doi:10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. PMID 10686599. S2CID 2916710.
- Hayashi S, McMahon AP (April 2002). "Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse". Developmental Biology. 244 (2): 305–18. doi:10.1006/dbio.2002.0597. PMID 11944939.
- Mouse genetics : methods and protocols. Singh, Shree Ram,, Coppola, Vincenzo. New York, NY. 26 July 2014. ISBN 9781493912155. OCLC 885338722.
{{cite book}}: CS1 maint: location missing publisher (link) CS1 maint: others (link) - Green JE, Ried T (2012). Genetically Engineered Mice for Cancer Research. doi:10.1007/978-0-387-69805-2. ISBN 978-0-387-69803-8. S2CID 40599715.
- Friedel RH, Wurst W, Wefers B, Kühn R (2011). "Generating Conditional Knockout Mice". Transgenic Mouse Methods and Protocols. Methods in Molecular Biology. Vol. 693. pp. 205–31. doi:10.1007/978-1-60761-974-1_12. ISBN 978-1-60761-973-4. PMID 21080282.
- Sakamoto K, Gurumurthy CB, Wagner K (2014), Singh SR, Coppola V (eds.), "Generation of Conditional Knockout Mice", Mouse Genetics, Methods in Molecular Biology, Springer New York, vol. 1194, pp. 21–35, doi:10.1007/978-1-4939-1215-5_2, ISBN 9781493912148, PMID 25064096
- Imuta Y, Kiyonari H, Jang CW, Behringer RR, Sasaki H (March 2013). "Generation of knock-in mice that express nuclear enhanced green fluorescent protein and tamoxifen-inducible Cre recombinase in the notochord from Foxa2 and T loci". Genesis. 51 (3): 210–8. doi:10.1002/dvg.22376. PMC 3632256. PMID 23359409.
- Hall B, Limaye A, Kulkarni AB (September 2009). "Overview: generation of gene knockout mice". Current Protocols in Cell Biology. Chapter 19: Unit 19.12 19.12.1–17. doi:10.1002/0471143030.cb1912s44. PMC 2782548. PMID 19731224.
- Rodrigues JV, Shakhnovich EI (2019-08-01). "Adaptation to mutational inactivation of an essential E. coli gene converges to an accessible suboptimal fitness peak". bioRxiv: 552240. doi:10.1101/552240.
- Schwenk F, Baron U, Rajewsky K (December 1995). "A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells". Nucleic Acids Research. 23 (24): 5080–1. doi:10.1093/nar/23.24.5080. PMC 307516. PMID 8559668.
- Griffiths AJ, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM (2000). "Inversions". An Introduction to Genetic Analysis. 7th Edition.
- Xu J, Zhu Y (August 2018). "A rapid in vitro method to flip back the double-floxed inverted open reading frame in a plasmid". BMC Biotechnology. 18 (1): 52. doi:10.1186/s12896-018-0462-x. PMC 6119287. PMID 30170595.
- Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K (November 2003). "Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71". Nucleic Acids Research. 31 (22): 140e–140. doi:10.1093/nar/gng140. PMC 275577. PMID 14602933.
- Xu J, Zhu Y (August 2018). "A rapid in vitro method to flip back the double-floxed inverted open reading frame in a plasmid". BMC Biotechnology. 18 (1): 52. doi:10.1186/s12896-018-0462-x. PMC 6119287. PMID 30170595.
- Griffiths AJ, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM (2000). "Translocations". An Introduction to Genetic Analysis (7th ed.).
- Pugach EK, Richmond PA, Azofeifa JG, Dowell RD, Leinwand LA (September 2015). "Prolonged Cre expression driven by the α-myosin heavy chain promoter can be cardiotoxic". Journal of Molecular and Cellular Cardiology. 86: 54–61. doi:10.1016/j.yjmcc.2015.06.019. PMC 4558343. PMID 26141530.
- Hayashi S, McMahon AP (April 2002). "Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse". Developmental Biology. 244 (2): 305–18. doi:10.1006/dbio.2002.0597. PMID 11944939.
- Hayashi S, McMahon AP (April 2002). "Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse". Developmental Biology. 244 (2): 305–18. doi:10.1006/dbio.2002.0597. PMID 11944939.
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP (December 1998). "Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase". Current Biology. 8 (24): 1323–6. doi:10.1016/s0960-9822(07)00562-3. PMID 9843687.
- Transgenesis techniques : principles and protocols. Clarke, Alan R. (2nd ed.). Totowa, NJ: Humana Press. 2002. ISBN 9781592591787. OCLC 50175106.
{{cite book}}: CS1 maint: others (link) - Cancer and zebrafish : mechanisms, techniques, and models. Langenau, David M. Switzerland. 10 May 2016. ISBN 9783319306544. OCLC 949668674.
{{cite book}}: CS1 maint: location missing publisher (link) CS1 maint: others (link) - Kobayashi K, Kamei Y, Kinoshita M, Czerny T, Tanaka M (January 2013). "A heat-inducible CRE/LOXP gene induction system in medaka". Genesis. 51 (1): 59–67. doi:10.1002/dvg.22348. PMID 23019184. S2CID 25211137.