Photolyase
Photolyases (EC 4.1.99.3) are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light (from the violet/blue end of the spectrum) both for their own activation[1] and for the actual DNA repair.[2] The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
| Cryptochrome/photolyase, C-terminal, FAD binding | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
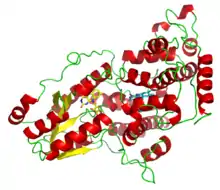 A deazaflavin photolyase from Anacystis nidulans, illustrating the two light-harvesting cofactors: FADH− (yellow) and 8-HDF (cyan). | |||||||||||
| Identifiers | |||||||||||
| Symbol | FAD_binding_7 | ||||||||||
| Pfam | PF03441 | ||||||||||
| InterPro | IPR005101 | ||||||||||
| PROSITE | PDOC00331 | ||||||||||
| SCOP2 | 1qnf / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| deoxyribodipyrimidine photo-lyase (CPD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 A UV radiation induced thymine-thymine cyclobutane dimer (right) is the type of DNA damage which is repaired by DNA photolyase. Note: The above diagram is incorrectly labelled as thymine as the structures lack 5-methyl groups. | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.99.3 | ||||||||
| CAS no. | 37290-70-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Function
Photolyases bind complementary DNA strands and break certain types of pyrimidine dimers that arise when a pair of thymine or cytosine bases on the same strand of DNA become covalently linked. The bond length of this dimerization is shorter than the bond length of normal B-DNA structure which produces an incorrect template for replication and transcription.[3] The more common covalent linkage involves the formation of a cyclobutane bridge. Photolyases have a high affinity for these lesions and reversibly bind and convert them back to the original bases.
Evolution
Photolyase is a phylogenetically old enzyme which is present and functional in many species, from the bacteria to the fungi to plants[4] and to the animals.[5] Photolyase is particularly important in repairing UV induced damage in plants. The photolyase mechanism is no longer working in humans and other placental mammals who instead rely on the less efficient nucleotide excision repair mechanism, although they do retain many cryptochromes.[6]
Photolyases are flavoproteins and contain two light-harvesting cofactors. Many photolyases have an N-terminal domain that binds a second cofactor. All photolyases contain the two-electron-reduced FADH−; they are divided into two main classes based on the second cofactor, which may be either the pterin methenyltetrahydrofolate (MTHF) in folate photolyases or the deazaflavin 8-hydroxy-7,8-didemethyl-5-deazariboflavin (8-HDF) in deazaflavin photolyases. Although only FAD is required for catalytic activity, the second cofactor significantly accelerates reaction rate in low-light conditions. The enzyme acts by electron transfer in which the reduced flavin FADH− is activated by light energy and acts as an electron donor to break the pyrimidine dimer.[7]
On the basis of sequence similarities DNA photolyases can be grouped into a few classes:[8][9]
|
- Class 1 CPD photolyases are enzymes that process cyclobutane pyrimidine dimer (CPD) lesions from Gram-negative and Gram-positive bacteria, the halophilic archaea Halobacterium halobium.
- Class 2 CPD photolyases also process CPD lesions. They are found in plants like the thale cress Arabidopsis thaliana and the rice.
- The plant and fungi cryptochromes are similar to Class 1 CPDs. They are blue light photoreceptors that mediate blue light-induced gene expression and modulation of circadian rhythms.
- Class 3 CPD lyases make up a sister group to the plant cryptochromes, which in turn are a sister group to class 1 CPDs.
- The Cry-DASH group are CPD lyases highly specific for single-stranded DNA. Members include Vibrio cholerae, X1Cry from Xenopus laevis, and AtCry3 from Arabidopsis thaliana.[5] DASH was initially named after Drosophila, Arabidopsis, Synechocystis, and Human, four taxa initially thought to carry this family of lyases. The categorization has since changed. The "Cry" part of their name was due to initial assumptions that they were cryptochromes.[8]
- Eukaryotic (6-4)DNA photolyases form a group with animal cryptochromes that control circadian rhythms. They are found in diverse species including Drosophila and humans. The cryptochromes have their own detailed grouping.[9]
- Bacterial 6-4 lyases (InterPro: IPR007357), also known as the FeS-BCP group, form their own outgroup relative to all photolyases.
The non-class 2 branch of CPDs tend to be grouped into class 1 in some systems such as PRINTS (PR00147). Although the members of the smaller groups are agreed upon, the phylogeny can vary greatly among authors due to differences in methodology, leading to some confusion with authors who try to fit everything (sparing FeS-BCP) into a two-class classification.[9] The cryptochromes form a polyphyletic group including photolyases that have lost their DNA repair activity and instead control circadian rhythms.[8][9]
Application
Adding photolyase from a blue-green algae Anacystis nidulans, to HeLa cells partially reduced DNA damage from UVB exposure.[10]
Nomenclature
The systematic name of this enzyme class is deoxyribocyclobutadipyrimidine pyrimidine-lyase. Other names in common use include photoreactivating enzyme, DNA photolyase, DNA-photoreactivating enzyme, DNA cyclobutane dipyrimidine photolyase, DNA photolyase, deoxyribonucleic photolyase, deoxyribodipyrimidine photolyase, photolyase, PRE, PhrB photolyase, deoxyribonucleic cyclobutane dipyrimidine photolyase, phr A photolyase, dipyrimidine photolyase (photosensitive), and deoxyribonucleate pyrimidine dimer lyase (photosensitive). This enzyme belongs to the family of lyases, specifically in the "catch-all" class of carbon-carbon lyases.
References
- Yamamoto J, Shimizu K, Kanda T, Hosokawa Y, Iwai S, Plaza P, Müller P (October 2017). "Loss of Fourth Electron-Transferring Tryptophan in Animal (6-4) Photolyase Impairs DNA Repair Activity in Bacterial Cells". Biochemistry. 56 (40): 5356–5364. doi:10.1021/acs.biochem.7b00366. PMID 28880077.
- Thiagarajan V, Byrdin M, Eker AP, Müller P, Brettel K (June 2011). "Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV". Proceedings of the National Academy of Sciences of the United States of America. 108 (23): 9402–7. Bibcode:2011PNAS..108.9402T. doi:10.1073/pnas.1101026108. PMC 3111307. PMID 21606324.
- Garrett RH, Grisham CM (2010). Biochemistry. Brooks/Cole, Cengage Learning. ISBN 978-0-495-10935-8. OCLC 984382855.
- Teranishi, M., Nakamura, K., Morioka, H.,Yamamoto, K. and Hidema, J. (2008). "The native cyclobutane pyrimidine dimer photolyase of rice is phosphorylated". Plant Physiology. 146 (4): 1941–1951. doi:10.1104/pp.107.110189. PMC 2287361. PMID 18235036.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Selby CP, Sancar A (November 2006). "A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity". Proceedings of the National Academy of Sciences of the United States of America. 103 (47): 17696–700. Bibcode:2006PNAS..10317696S. doi:10.1073/pnas.0607993103. PMC 1621107. PMID 17062752.
- Lucas-Lledó JI, Lynch M (May 2009). "Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family". Molecular Biology and Evolution. 26 (5): 1143–53. doi:10.1093/molbev/msp029. PMC 2668831. PMID 19228922.
- Sancar A (June 2003). "Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors". Chemical Reviews. 103 (6): 2203–37. doi:10.1021/cr0204348. PMID 12797829.
- Scheerer P, Zhang F, Kalms J, von Stetten D, Krauß N, Oberpichler I, Lamparter T (May 2015). "The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways". The Journal of Biological Chemistry. 290 (18): 11504–14. doi:10.1074/jbc.M115.637868. PMC 4416854. PMID 25784552.
- Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, Oakley TH (April 2012). "Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin". The Journal of Experimental Biology. 215 (Pt 8): 1278–86. doi:10.1242/jeb.067140. PMC 3309880. PMID 22442365.
- Kulms D, Pöppelmann B, Yarosh D, Luger TA, Krutmann J, Schwarz T (July 1999). "Nuclear and cell membrane effects contribute independently to the induction of apoptosis in human cells exposed to UVB radiation". Proceedings of the National Academy of Sciences of the United States of America. 96 (14): 7974–9. Bibcode:1999PNAS...96.7974K. doi:10.1073/pnas.96.14.7974. PMC 22172. PMID 10393932.
Further reading
- Eker AP, Fichtinger-Schepman AM (1975). "Studies on a DNA photoreactivating enzyme from Streptomyces griseus II. Purification of the enzyme". Biochim. Biophys. Acta. 378 (1): 54–63. doi:10.1016/0005-2787(75)90136-7. PMID 804322.
- Sancar GB, Smith FW, Reid R, Payne G, Levy M, Sancar A (1987). "Action mechanism of Escherichia coli DNA photolyase. I. Formation of the enzyme-substrate complex". J. Biol. Chem. 262 (1): 478–85. doi:10.1016/S0021-9258(19)75952-3. PMID 3539939.
- Setlow JK, Bollum FJ (1968). "The minimum size of the substrate for yeast photoreactivating enzyme". Biochim. Biophys. Acta. 157 (2): 233–7. doi:10.1016/0005-2787(68)90077-4. PMID 5649902.
External links
 Media related to Photolyase at Wikimedia Commons
Media related to Photolyase at Wikimedia Commons