Dendroscope
Dendroscope is an interactive computer software program written in Java for viewing Phylogenetic trees.[1] This program is designed to view trees of all sizes and is very useful for creating figures. Dendroscope can be used for a variety of analyses of molecular data sets but is particularly designed for metagenomics or analyses of uncultured environmental samples.
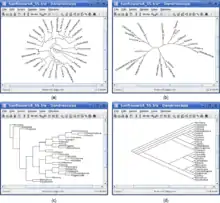 Dendroscope: (a) circular cladogram, (b) radial phylogram, (c) rectangular phylogram, and (d) slanted cladogram. Image by Huson et al. | |
| Developer(s) | Daniel Huson et al. |
|---|---|
| Stable release | 3.6.0
/ 2019 |
| Repository | |
| Operating system | Windows, Linux, Mac OS X |
| Type | Bioinformatics |
| License | GPLv3 or later |
| Website | http://ab.inf.uni-tuebingen.de/software/dendroscope/ |
It was developed by Daniel Huson and his colleagues at the University of Tübingen in Germany, who also created SplitsTree.
References
- Huson, Daniel H.; Daniel C. Richter; Christian Rausch; Tobias Dezulian; Markus Franz; Regula Rupp (2007-11-22). "Dendroscope: An interactive viewer for large phylogenetic trees". BMC Bioinformatics. United Kingdom: BioMedCentral. 8: 460. doi:10.1186/1471-2105-8-460. PMC 2216043. PMID 18034891.
External links
- Dendroscope homepage
- List of phylogeny software, hosted at the University of Washington
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.