< GENtle
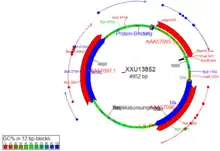

The DNA map.
The DNA map is shown for DNA sequences (though a variant is also used in protein module for the schematics display). It shows the linear or circular (e.g., plasmid) DNA sequence as a map.
Display
Displayed are
- features (including optional sequencing primers)
- restriction sites
- sticky ends (if any)
- open reading frames (optional)
- sequence name and length (optional, see Options)
- methylation (optional, see Options)
- GC contents (optional, see Options)
Mouse actions
| Action on | Mouse button | Function |
|---|---|---|
| Background | left | Mark sequence |
| left (double click) | Open Sequence editor | |
| middle | Show marked DNA in sequence; show current position in sequence if nothing is marked | |
| Feature | left | Move feature display |
| left (double click) | Edit feature (see Sequence editor) | |
| middle | Mark DNA that matches feature | |
| middle (shift pressed) | Extend currently marked area to include the DNA of the feature | |
| Restriction site | left | Move site display |
| left (double click) | Edit enzyme list (see Sequence editor) | |
| middle | Open Restriction Assistant | |
| Open reading frame | left | Mark ORF sequence |
| left (double click) | Mark and show ORF sequence | |
| All | right | Context menu |
Context menu
The context menu opens on a click with the right mouse button when somewhere inside the DNA map. The contents of the menu depends on what object in the map you clicked on. Also, depending on the properties of the object, some functions might not be available, for example, amino acids of a feature with no reading frame.
Background
| Edit sequence | Opens the Sequence editor |
| Transform sequence | Make sequence inverted and/or complementary |
| Limit enzymes | Limits enzymes to those that cut no more than n times |
| PCR/PCR | Starts the PCR modul |
| PCR/Forward | Starts the PCR modul and generates a 5'->3'-primer |
| PCR/Backward | Starts the PCR modul and generates a 5'->3'-primer |
| PCR/Both | Starts the PCR modul and generates both primers |
| PCR/Mutation | Starts the PCR modul and generates overlapping mutagenesis primers |
| Selection/Cut | Removes the selected part of the sequence and puts it into the clipboard |
| Selection/Copy | Copys the selected part of the sequence into the clipboard |
| Selection/Copy to new sequence | Generate a new DNA sequence entry based on the selection |
| Selection/Show enzymes that cut here | Opens a variant of the Silent Mutagenesis dialog for the selected part of the sequence |
| Selection/Selection as new feature | Generates a new feature for the selected part of the sequence |
| Selection/Extract amino acids | Extracts the amino acid sequence of the selected part of the DNA sequence |
| Selection/BLAST amino acids | Runs a BLAST search for the amino acid sequence of the selected part of the DNA sequence |
| Selection/BLAST DNA | Runs a BLAST search for the selected part of the DNA sequence |
| Sequence map/Save as image | Saves the DNA map as an image file |
| Sequence map/Copy image to clipboard | Copies the DNA map as a bitmap or WMF (see Options) to the clipboard |
| Sequence map/Print map | Prints the DNA map |
| Show/hide ORFs | Toggles the open reading frame display |
| Edit ORFs | Adjusts the open reading frame display |
Restriction sites
| Edit restriction enzyme | Add/remove/manage restriction enzyme via the Sequence editor |
| Show/hide enzyme | Toggle visibility for the enzyme (this will affect all restriction sites for that enzyme in this sequence) |
| Remove enzyme | Remove the enzyme from the current selection (this will affect all restriction sites for that enzyme in this sequence). This will not work for automatically added enzymes (see Options) |
| Mark restriction site | Marks the recognition sequence of that enzyme at that restriction site |
| Mark and show restriction site | Marks the recognition sequence of that enzyme at that restriction site and shows it in the sequence |
| Online enzyme information | Opens the ReBase page for that enzyme |
| Add to cocktail | This adds the enzyme to the restriction cocktail (see Restriction Assistant) |
| Add to cocktail | This adds the enzyme to the restriction cocktail (see Restriction Assistant) and starts the restriction |
Features
| Edit feature | Edit the feature properties (see GENtle/Sequence_editor) |
| Hide feature | Hide the feature from display |
| Delete feature | Delete the feature |
| DNA Sequence/Mark feature sequence | Mark the DNA sequence that matches the feature |
| DNA Sequence/Mark and show feature sequence | Mark the DNA sequence that matches the feature and shows it in the sequence |
| DNA Sequence/Copy (coding) DNA sequence | Copies the DNA sequence that matches the feature to the clipboard |
| DNA Sequence/This feature as new sequence | Generates a new DNA sequence based on the feature |
| DNA Sequence/BLAST DNA | Runs a BLAST search for the DNA of the feature |
| Amino acid sequence/Copy amino acid sequence | Copies the amino acid sequence of the feature to the clipboard |
| Amino acid sequence/As new entry | Generates a new protein entry based on the amino acid sequence of the feature |
| Amino acid sequence/Blast amino acids | Runs a BLAST search for the amino acid sequence of the feature |
Open reading frames
| As new feature | Generate a new feature from the ORF, with the appropriate reading frame and direction |
| DNA sequence/Copy DNA sequence | Copies the DNA sequence of the ORF to the clipboard |
| DNA sequence/As new DNA | Generates a new DNA sequence entry based on the DNA sequence of the ORF |
| DNA sequence/BLAST DNA | Runs a BLAST search for the DNA sequence of the ORF |
| Amino acid sequence/Copy amino acid sequence | Copies the amino acid sequence of the ORF to the clipboard |
| Amino acid sequence/As new AA | Generates a new protein entry based on the amino acid sequence of the ORF |
| Amino acid sequence/BLAST amino acids | Runs a BLAST search for the amino acid sequence of the ORF |
This article is issued from Wikibooks. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.